Research Article - Journal of Drug and Alcohol Research ( 2022) Volume 11, Issue 9
Pyridine moiety bearing pyrimidine-2-thiols: Synthesis, characterization and insilico molecular docking studies in comparison to standard ligand drug
K. Neelaveni1* and Y. Rajendra Prasad22Department of Pharmaceutical Sciences, AU College of Pharmaceutical Sciences, India
K. Neelaveni, Department of Pharmaceutical Sciences, Aacharya Nagarjuna University, India, Email: neelaveni210@gmail.com
Received: 31-Aug-2022, Manuscript No. jdar-22-77918; Editor assigned: 02-Sep-2022, Pre QC No. jdar-22-77918 (PQ); Reviewed: 16-Sep-2022, QC No. jdar-22-77918; Revised: 21-Sep-2022, Manuscript No. jdar-22-77918 (R); Published: 28-Sep-2022, DOI: 10.4303/jdar/236197
Abstract
Using specified synthesis process, chalcones were converted into a new pyridine moiety containing pyrimidine-2-thiols. By combining 3-acetylpyridine with substituted benzaldehydes, pyridine-bearing chalcones were developed initialy. New pyrimidine-2-thiol derivatives are produced by the reaction of additional pyridine containing chalcones with thiourea. Physical and spectral analysis was used to describe the derivatives of pyridine containing pyrimidine-2-thiol that were generated. When compared to standard ligand drugs, 4-(4-nitrophenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol exhibits significant binding interaction at the active site regions of cyclooxygenase-1 and cyclooxygenase-2, according to molecular docking studies performed at cyclooxygenase targets using Auto-Dock Vina software.
Keywords
3-acetylpyridine; Substituted Benzaldehydes; Chalcones; Pyrimidine- 2-Thiols; Molecular Docking
Introduction
A well-known starting point for the synthesis of several heterocyclic compounds is the chalcone scaffold. Due to their potential for application in medicine, pyrazolines and pyrimidines, which are heterocyclic compounds with nitrogen-containing rings, have become more popular as a result of chalcone cyclization [1-3]. Due to their wide range of biological activity, heterocyclic compounds, especially those with five or six members to the ring, have dominated other families of organic molecules. From the time of their discovery as crucial components of nucleic acids to the present, pyrimidines have a long and illustrious history. They are currently used in chemotherapy. As a crucial component of all cells and consequently of all living things, pyrimidines are one of the most significant heterocyclic compounds among all and exhibit exceptional phar macological actions. Numerous simple fused pyrimidines, such as purines and pteridines, are biologically active on their own and are crucial building blocks of highly significant naturally occurring compounds. One of the potential explanations for their activity is that the fundamental components of nucleic acids (DNA and RNA), cytosine, uracil, and thymine, all include pyrimidine bases. Pyrimidines are significant classes of heterocyclic compounds and have a wide range of biological effects, including anti-inflammatory, analgesic, antibacterial, anti-HIV, anti-allergic, anti-malerial, and anticancer effects [4-7]. This study concentrated on synthesizing pyridine-containing pyrimidine scaffolds from chalcones. Claisen-Schmidt condensation was used to create chalcone chemicals. Chalcones and thiourea underwent cyclization to produce a variety of pyrimidine-2-thiol derivatives. Physical and spectroscopic characteristics were used to categorise newly synthesised pyridine containing pyrimidine-2-thiol derivatives. The active site regions of cyclooxegenase-1 (PDB ID: 3KK6) and cyclooxegenase-2 (PDB ID: 5IKR) were studied using insilico molecular docking studies to estimate anti-inflammatory effects in contrast to conventional ligands.
Materials and Methods
Commercial suppliers Merck and AVRA provided the chemicals and reagents that were employed for the synthesis of novel pyrimidine-2-thiol derivatives; these materials were not purified before use. With the use of E.Merck grade silica gel 60GF-254 pre-coated plates, thin layer chromatography was used to monitor both the reaction’s progress and completion. Uncorrected electrical melting point apparatus was used to determine melting points. IR spectra [KBr in cm-1] of the substances were recorded using the KBr pellet technique in a Bruker FT-IR spectrophotometer. On a Bruker-AMX spectrophotometer operating at 400 MHz, chemical shifts in ppm of 1H-NMR spectra were noted in respect to tetra methyl silane (TMS) as the internal standard. Using an Agilent LC-MSD-1200 mass spectrometer, the mass spectral data were recorded. Using Auto-DockVina, ChemDraw, and BIOVIA discovery studio software, molecular docking studies were performed at several target protein active sites to investigate binding interactions in contrast with standard ligand.
Synthesis of pyridine bearing chalcone derivatives 3a- 3h
Claisen-Schmidt condensation of 3-acetylpyridine 1 (0.01 mol) with substituted benzaldehyde 2 (0.01 mol) in 20 ml of ethanol in the presence of 20% NaOH as a catalyst. The reaction mixture was stirred at room temperature for 6–18 hrs. Progress of the reaction was monitored by TLC. The precipitate formed was filtered, washed with cold water, and dried to give crude chalcones. The crude solid chalcone products were recrystallized from methanol afford chalcones [8,9].
Synthesis of pyridine bearing pyrimidine-2-thiol derivatives from chalcones 4a-4h
The preparation of pyrimidine derivatives involves the reaction of chalcones 3a-3h with thiourea. A mixture of chalcone (0.01 mol), thiourea (0.01 mol), ethanolic NaOH (5 g of NaOH in 25 ml of ethanol) was refluxed for 12-15 hrs. The reaction progress was monitored by TLC. The precipitate formed was filtered, washed with ice cold water, and dried. The crude solid product was recrystallized from ethanol [10-13].
4a: 4-(4-nitrophenyl)-6-(pyridin-3-yl)-pyrimidine-2-thiol
IR [KBr ѵ cm-1]: 2568 (pyrimidine-SH str.), 3056 (aromatic= C-H str.), 1558 (aromatic C=C str.), 1652 (C=N str.), 1538 and 1357 (nitro N-O str.). 1H-NMR [400 MHz, δ, ppm, DMSO-d6]: 1.93 (1H, s, -SH), 8.49 (1H, s, pyrimidine- C5-H), 7.06-7.18 (2H, d, phenyl-C2-H and C6- H),8.05-8.17 (2H, d, phenyl-C3-H and C5-H), 8.85 (1H, s, pyridine-C2-H), 8.29-8.37 (1H, d, pyridine-C4-H), 7.45-7.60 (1H, t, pyridine-C5-H), 8.50-8.63 (1H, d, pyridine- C6-H). ESI-MS: m/z (M+) 310.05
4b: 4-(4-chlorophenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol
IR [KBr ѵ cm-1]: 2571 (pyrimidine-SH str.), 3045 (aromatic= C-H str.), 1572 (aromatic C=C str.), 1659 (C=N str.), 786 (C-Cl str.). 1H-NMR [400 MHz, δ, ppm, DMSO- d6]: 1.78 (1H, s, -SH), 8.53 (1H, s, pyrimidine-C5-H), 7.78-7.90 (2H, d, phenyl-C2-H and C6-H),8.23-8.35 (2H, d, phenyl-C3-H and C5-H), 8.96 (1H, s, pyridine-C2-H), 8.33-8.46 (1H, d, pyridine-C4-H), 7.52-7.74 (1H, t, pyridine- C5-H), 8.67-8.79 (1H, d, pyridine-C6-H).ESI-MS: m/z (M+) 299.03
4c: 4-(2-chlorophenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol
IR [KBr ѵ cm-1]: 2559 (pyrimidine-SH str.), 3073 (aromatic= C-H str.), 1498 (aromatic C=C str.), 1638 (C=N str.), 798 (C-Cl str.). 1H-NMR [400 MHz, δ, ppm, DMSO- d6]: 1.91 (1H, s, -SH), 8.41 (1H, s, pyrimidine-C5-H), 7.35-7.81 (4H, m, phenyl-H), 8.90 (1H, s, pyridine-C2-H), 8.21-8.34 (1H, d, pyridine-C4-H), 7.40-7.62 (1H, t, pyridine- C5-H), 8.58-8.70 (1H, d, pyridine-C6-H).ESI-MS: m/z (M+) 299.03
4d: 4-(4-bromophenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol
IR [KBr ѵ cm-1]: 2563 (pyrimidine-SH str.), 3055 (aromatic= C-H str.), 1520 (aromatic C=C str.), 1643 (C=N str.), 593 (C-Brstr.). 1H-NMR [400 MHz, δ, ppm, DMSO- d6]: 1.69 (1H, s, -SH), 8.66 (1H, s, pyrimidine-C5-H), 7.68-7.80 (2H, d, phenyl-C2-H and C6-H),7.98-8.10 (2H, d, phenyl-C3-H and C5-H), 8.85 (1H, s, pyridine-C2-H), 8.31-8.43 (1H, d, pyridine-C4-H), 7.41-7.63 (1H, t, pyridine- C5-H), 8.61-8.73 (1H, d, pyridine-C6-H).ESI-MS: m/z (M+) 342.98
4e: 4-(4-hydroxyphenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol
IR [KBr ѵ cm-1]: 2577 (pyrimidine-SH str.), 3069 (aromatic= C-H str.), 1485 (aromatic C=C str.), 1663 (C=N str.), 3559 (phenolic-OH str.). 1H-NMR [400 MHz, δ, ppm, DMSO- d6]: 1.85 (1H, s, -SH), 5.34 (1H, s, phenolic-OH), 8.58 (1H, s, pyrimidine-C5-H), 7.59-7.70 (2H, d, phenyl-C2-H and C6-H),6.91-7.04 (2H, d, phenyl-C3-H and C5-H), 8.71 (1H, s, pyridine-C2-H), 8.25-8.37 (1H, d, pyridine-C4-H), 7.33-7.56 (1H, t, pyridine-C5-H), 8.59-8.71 (1H, d, pyridine- C6-H).ESI-MS: m/z (M+) 281.06 4f: 4-(4-methylphenyl)-6-(pyridin-3-yl
4f: 4-(4-methylphenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol
IR [KBr ѵ cm-1]: 2560 (pyrimidine-SH str.), 3081 (aromatic= C-H str.), 1524 (aromatic C=C str.), 1656 (C=N str.), 2937 (-CH str.). 1H-NMR [400 MHz, δ, ppm, DMSO- d6]: 1.98 (1H, s, -SH), 2.35 (3H, s, -CH3), 8.66 (1H, s, pyrimidine-C5-H), 7.61-7.74 (2H, d, phenyl-C2-H and C6-H), 6.95-7.08 (2H, d, phenyl-C3-H and C5-H), 8.82 (1H, s, pyridine-C2-H), 8.31-8.44 (1H, d, pyridine-C4-H), 7.43-7.65 (1H, t, pyridine-C5-H), 8.41-8.52 (1H, d, pyridine- C6-H).ESI-MS: m/z (M+) 279.08
4g: 4-(4-methoxyphenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol
IR [KBr ѵ cm-1]: 2582 (pyrimidine-SH str.), 3047 (aromatic= C-H str.), 1551 (aromatic C=C str.), 1660 (C=N str.), 1156 (ether C-O str.), 2916 (-CH str.). 1H-NMR [400 MHz, δ, ppm, DMSO-d6]: 1.53 (1H, s, -SH), 3.83 (3H, s, -OCH3), 8.75 (1H, s, pyrimidine-C5-H), 7.71-7.82 (2H, d, phenyl-C2-H and C6-H),7.05-7.18 (2H, d, phenyl-C3-H and C5-H), 8.79 (1H, s, pyridine-C2-H), 8.29-8.41 (1H, d, pyridine-C4-H), 7.51-7.72 (1H, t, pyridine-C5-H), 8.64- 8.76 (1H, d, pyridine-C6-H).ESI-MS: m/z (M+) 295.08
4h: 4-(4-hydroxy-3-methoxy-phenyl)-6-(pyridin-3-yl)- pyrimidine-2-thiol
IR [KBr ѵ cm-1]: 2567 (pyrimidine-SH str.), 3573 (phenolic- OH str.), 3088 (aromatic=C-H str.), 1498 (aromatic C=C str.), 1643 (C=N str.), 1185 (ether C-O str.), 2889 (-CH str.). 1H-NMR [400 MHz, δ, ppm, DMSO-d6]: 1.86 (1H, s, -SH), 3.88 (3H, s, -OCH3), 5.27 (1H, s, phenolic- OH), 8.68 (1H, s, pyrimidine-C5-H), 7.20 (1H, d, phenyl-C2-H),6.90-7.04 (1H, d, phenyl-C5-H), 7.25- 7.36 (1H, d, phenyl-C6-H),8.66 (1H, s, pyridine-C2-H), 8.31-8.42 (1H, d, pyridine-C4-H), 7.68-7.89 (1H, t, pyridine- C5-H), 8.05-8.18 (1H, d, pyridine-C6-H).ESI-MS: m/z (M+) 311.07
Molecular docking studies
In order to predict the molecular mechanisms at the target proteins of cyclooxygenase-1 (COX-1) and cyclooxygenas- 2 (COX-2) as possible anti-inflammatory drugs, molecular docking experiments were conducted. Downloadable PDB files for the 3D crystal structures of cyclooxegenase- 1 PDB ID: 3KK6 [14-17] and cyclooxegenase-2 PDB ID: 5IKR [18-21] are available from the RCBS Protein Data Bank at (https://www.rcsb.org/). Using BIOVIA Discovery studio visualize 2021, the recovered proteins were made ready for docking by having water molecules and hetero atoms removed. By adding Kollman charges, Gasteiger charges, and polar hydrogens, the protein structures were reduced to the lowest energy state for further investigation. ChemDraw Ultra 12.0 was used to create the intended and produced ligand structures, and Chem 3D-Pro 12.0 was used to reduce and store the structures in SDF file format. From the Pubchem database, the Diclofenac standard ligand structure SDF file formats (Pubchem Id: 3033) were downloaded. Additionally, using the Open Babel software, all SDF format files are translated to PDB format. Protein transformations from PDB to PDBQT file format, grid-based docking studies using default parameters, and docking by connecting the protein-ligand using MGL Auto-Dock Vina were all completed. Using command prompt, it was possible to see the docking positions of ligands that best linked to the active site areas of the proteins. The bindings of the ligands to the target proteins in two and three dimensions were viewed using BIOVIA Discovery Studio.
Results and Discussion
Initially pyridine bearing chalcones (3a-3h) were synthesized by the Claisen-Schmidt reaction of 3-acetylpyridine and substituted benzaldehydes. Further, pyridine bearing chalcones treated with thiourea undergoes cyclization to afford pyridine bearing pyrimidine-2-thiol derivatives (4a- 4h). The scheme of synthesis of titled pyrimidine-2-thiol derivatives was depicted in Figures 1 and 2 and respective compounds physical characterization data was shown in Table 1.
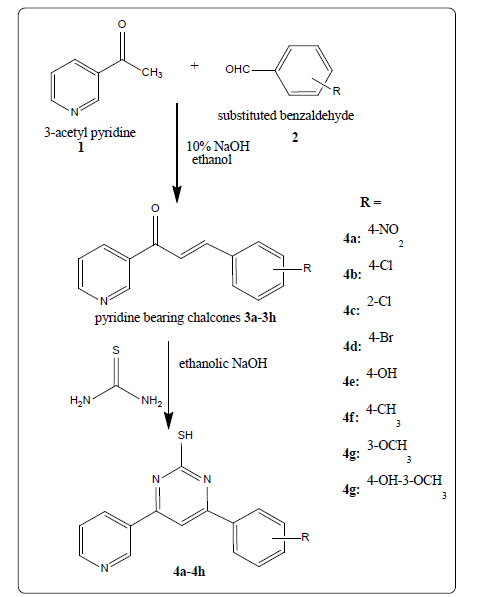
Figure 1: Pyrimidine-2-thiol derivatives synthesis from chalcones
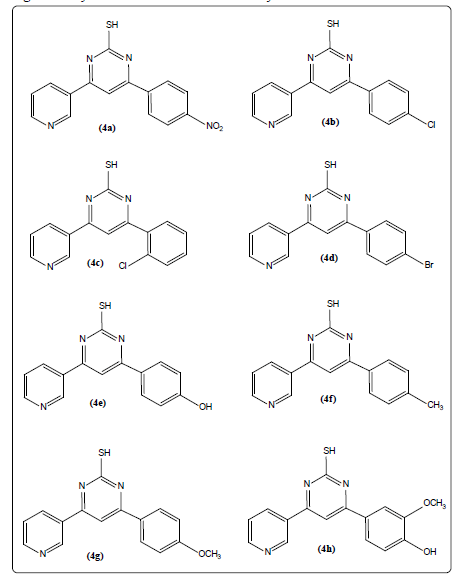
Figure 2: Library of pyrimidine-2-thiol derivatives developed
| Compd. | R | m.p. (ºC) | Molecular formula | mol. wt. | % yield | reaction time | Rf value |
|---|---|---|---|---|---|---|---|
| 4a | 4-nitro | 172-174 | C15H10N4O2S | 310.33 | 75.64 | 13.5 hrs | 0.58 |
| 4b | 4-chloro | 180-182 | C15H10ClN3S | 299.78 | 69.35 | 14 hrs | 0.61 |
| 4c | 2-chloro | 168-170 | C15H10ClN3S | 299.78 | 72.25 | 12.5 hrs | 0.52 |
| 4d | 4-bromo | 160-162 | C15H10BrN3S | 344.23 | 70.48 | 14.5 hrs | 0.56 |
| 4e | 4-hydroxy | 170-172 | C15H11N3OS | 281.33 | 67.68 | 13 hrs | 0.60 |
| 4f | 4-methyl | 178-180 | C16H13N3S | 279.36 | 74.90 | 13.5 hrs | 0.59 |
| 4g | 4-methoxy | 166-168 | C16H13N3OS | 295.36 | 75.48 | 14 hrs | 0.58 |
| 4h | 4-hydroxy-3-methoxy | 164-166 | C16H13N3O2S | 311.36 | 72.56 | 13.5 hrs | 0.62 |
Table 1: Physical characterization data of pyrimidine-2-thiol derivatives 4a-4h
Molecular docking results
Anti-inflammatory activities have been predicted computationally by performing molecular docking studies at cyclooxygenase- 1 and cyclooxygenase-2 targets.
At cyclooxygenase-1 PDB ID: 3KK6: In comparison to the conventional ligand Diclofenac, designed ligands were evaluated in the cyclooxygenase-1 active site area (3KK6). Table 2, Figures 3 and 4 demonstrate the outcomes of screening for the cyclooxygenase-1 protein 3KK6 using designed ligands and the common ligand Diclofenac. The common ligand Diclofenac was shown to have a lower docking energy −4.20 kcal/mol at the active site region than the developed ligand 4a docking energy −4.72 kcal/ mol, which suggests that 4a shown significantly more likely to bind to the cyclooxygenase-1 protein.
| Compound | Binding energy (kcal/mol) | Interacted amino acid residues |
|---|---|---|
| Diclofenac | -4.20 | ALA133, PHE220, ILE132 |
| 4a | -4.72 | ILE165, HIS215, LEU240, MET198, ALA152 |
| 4b | -4.08 | MET216, ALA133, ILE132, SER146, ASN144 |
| 4c | -3.97 | SER232, TYR210, ARG185, ILE155 |
| 4d | -3.38 | PHE220, ASN156, CYS252, MET241, ALA134 |
| 4e | -4.02 | HIS167, ASN184, ILE160, TYR255 |
| 4f | -3.10 | GLY222, GLN268, PHE220, CYS180, HIS153 |
| 4g | -3.84 | GLN205, MET176, SER164, ILE266, ARG285 |
| 4h | -3.46 | CYS280, TYR261, ILE175, MET251, ARG193 |
Table 2: Binding energy and amino acid residues interacted with COX-1 protein target PDB ID–3KK6.
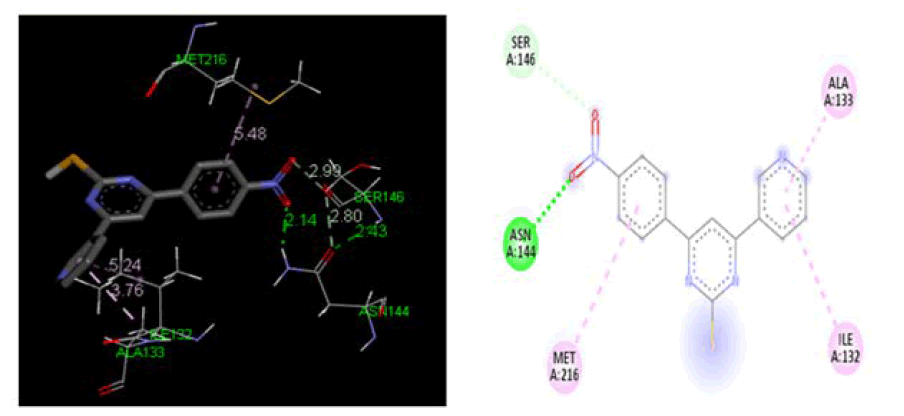
Figure 3: Three-dimensional& two-dimensional binding mode of designed ligand compounds 4a at active site region of cyclooxygenase-2 protein PDB ID- 3KK6
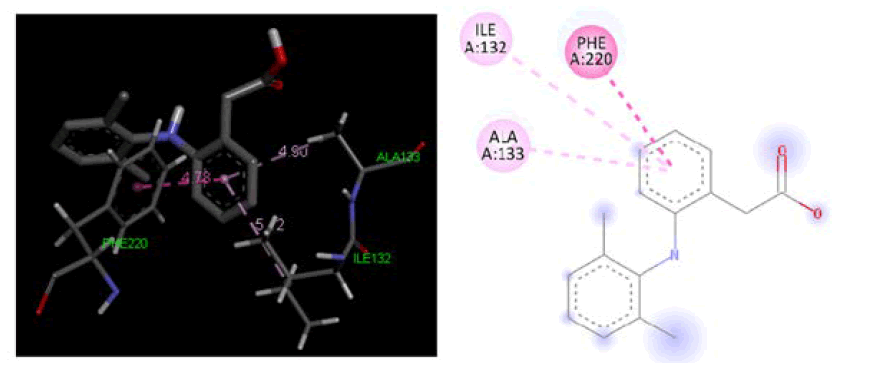
Figure 4: Three-dimensional& two-dimensional binding mode of standard ligand Diclofenac at active site region of cyclooxygenase-1 protein PDB ID- 3KK6
At cyclooxygenase-2PDB ID: 5IKR: In comparison to the common ligand Diclofenac, the designed ligand was tested in the cyclooxygenase-2 active site area (5IKR). Table 3, Figures 5 and 6 as well as the results of the screening for the cyclooxygenase-2 protein 5IKR using designed ligands and the reference ligand Diclofenac, were displayed. A much more likely ligand at the cyclooxygenase-2 protein was revealed to be compound 4a, whose docking energy at the active site region was reported to be −4.90 kcal/mol greater than that of reference ligand Diclofenac docking energy −4.28 kcal/mol.
| Compound | Binding energy (kcal/mol) | Interacted amino acid residues |
|---|---|---|
| Diclofenac | -4.28 | GLY217, PRO218, ALA132, ASP133, ALA219 |
| 4a | -4.90 | ILE150, GLN359, ASN299, ARG163 |
| 4b | -3.96 | ARG216, ASP133, ALA219, GLN457, ALA132 |
| 4c | -3.82 | PHE239, HIS228, CYS175, ILE154, SER238 |
| 4d | -2.97 | ALA218, MET245, PHE195, ARG252 |
| 4e | -3.49 | HIS198, SER208, ALA148, ILE220 |
| 4f | -2.76 | ASP165, ASN248, PRO195, MET218, CYS164 |
| 4g | -3.68 | LEU232, GLN347, ASN186, ARG186, PHE215 |
| 4h | -3.07 | ILE225, ASP162, SER235, HIS227, ALA194 |
Table 3: Binding energy and amino acid residues interacted with COX-2 protein target PDB ID–5IKR.
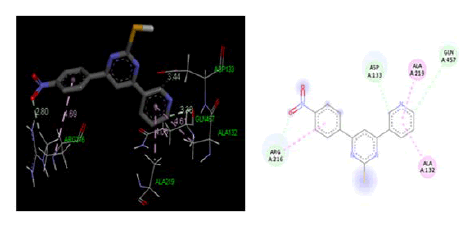
Figure 5: Three-dimensional& two-dimensional binding mode of designed ligand compounds 4a at active site region of cycloxygenase-2 protein PDB ID- 5IKR
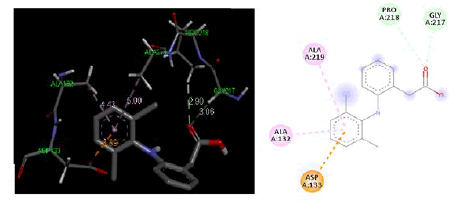
Figure 6: Three-dimensional & two-dimensional binding mode of standard ligand Diclofenac at active site region of cyclooxygenase-2 protein-PDB ID- 5IKR
Conclusion
Various novel pyrimidine-2-thiol derivatives 4a–4h containing pyridine moiety were developed and produced as named compounds in this investigation. The chemicals were physically and spectrally characterized. Through computational screening at the COX-1 (PDB ID-3KK6) and COX-2 (PDB ID-5IKR) receptor proteins, all the compounds were tested for their anti-inflammatory properties. When compared to the standard ligand Diclofenac, compound 4a, 4-(4-nitrophenyl)-6-(pyridin-3-yl)-pyrimidine- 2-thiol, exhibits significant binding interaction at the active site regions of the target protein, indicating that 4a is the leading candidate for the development of efficient and secure anti-inflammatory drugs.
Acknowledgements
The authors are thankful to the Management and staff of CMR College of Pharmacy, Hyderabad, Telangana, India for providing necessary facilities to carry out the research work
Conflict of Interest
The authors declare that they do not have any conflict of interest related to the matter or content discussed in this original research article
References
- S. Sinha, B. Medhi, R. Sehgal, Chalcones as an emerging lead molecule for antimalarial therapy: A review, J Mdrn Med Chem, 1(2013):64–77.
[Crossref] [Google Scholar] [PubMed]
- M.M. Alidmat, M. Khairuddean, N.M. Norman, A.N. Asri, M.H. Suhaimi, Synthesis, characterization, docking study and biological evaluation of new chalcone, pyrazoline, and pyrimidine derivatives as potent antimalarial compounds, Arbn J Chem, 14(2021):103304.
[Crossref] [Google Scholar] [PubMed]
- K.S. Jain, N. Arya, N.N. Inamdar, P.B. Auti, S.A. Unawane, et al. The chemistry and bio-medicinal significance of pyrimidines & condensed pyrimidines, Curr Tpcs in Med Chem, 16(2016):3133–3174.
[Crossref] [Google Scholar] [PubMed]
- K. Nagaraju, G. Lalitha, M. Suresh, G. Kranthi Kumar, B.J. Sreekantha, A review on recent advances in nitrogen-containing molecules and their biological applications, J Molecules, 25(2020):1909.
[Crossref] [Google Scholar] [PubMed]
- M. Satish Kumar, M. Vijey Aanandhi, An insight into the therapeutic potential of pyridopyrimidines as anticancer agents, Res J Pharm Tech, 11(2018):1259-1269.
[Crossref] [Google Scholar] [PubMed]
- S. Kumar, B. Narasimhan, Therapeutic potential of heterocyclic pyrimidine scaffolds, J Chem Cen, 12(2018).
[Crossref] [Google Scholar] [PubMed]
- H.M. Ashour, O.G. Shaaban, O.H. Rizk, I.M. El-Ashmawy, Synthesis and biological evaluation of thieno [2′,3′:4,5] pyrimido[1,2-b][1,2,4]-triazines and thieno[2,3-d] [1,2,4]triazolo[1,5-a]pyrimidines as anti-infammatory and analgesic agents, Eur J Med Chem, 62(2013):341–351.
[Crossref] [Google Scholar] [PubMed]
- M.M. Ibrahim, M. Al-Refai, R. Abu El-Halawa, Synthesis of some new chalcone and 4, 5-dihydro-1H-pyrazole derivatives as potential antimicrobial agents, Jordan J Chem, 146(2012):1–9.
[Crossref] [Google Scholar] [PubMed]
- K.A. Salum, M.M. Alidmat, M. Khairulddean, N.N.S.N.M. Kamal, M. Muhammad, Design, synthesis, characterization, and cytotoxicity activity evaluation of mono-chalcones and new pyrazolines derivatives, J Appl Pharm Sci, 10(2020):20–36.
[Crossref] [Google Scholar] [PubMed]
- M.M.M. Ramiz, W.A. El-Sayed, A.I. El-Tantawy, A.A. Abdel-Rahman, Antimicrobial activity of new 4,6-disubstituted pyrimidine, pyrazoline, and pyran derivatives, Arch Pharm Res, 33(2010):647-654.
[Crossref] [Google Scholar] [PubMed]
- A. Ahmed, A.l. Hutham, W. Ruaa, Synthesis, characterization, and antibacterial activity of some new pyrimidine derivatives from chalcone derivatives, Drug Inv Tdy, 11(2019):1732-1739.
[Crossref] [Google Scholar] [PubMed]
- B.M. Sahoo, M. Rajeswari, P. Jnyanaranjan, S. Binayani, Green expedient synthesis of pyrimidine derivatives via chalcones and evaluation of their anthelmintic activity, Indn J Pharm Edu Res, 51(2017):S700-S706.
[Crossref] [Google Scholar] [PubMed]
- M.S.K. Youssef, A.A.O. Abeed, Synthesis, characterization and pharmacological activities of pyrimidine derivatives containing 2-pyrazoline-5-one, Int J Pharm Sci Res, 5(2014):1705-1720.
[Crossref] [Google Scholar] [PubMed]
- G. Lauro, M. Manfra, S. Pedatella, K. Fischer, V. Cantone, et al. Identification of novel microsomal prostaglandin E2 synthase-1 (mPGES-1) lead inhibitors from fragment virtual screening, Eur J Med Chem, 125(2017):278−287.
[Crossref] [Google Scholar] [PubMed]
- F. Rorsch, E. Buscato, K. Deckmann, G. Schneider, M. Schubert-Zsilavecz, et al. Structure−activity relationship of nonacidic quinazolinone inhibitors of human microsomal prostaglandin synthase-1 (mPGES-1), J Med Chem, 55(2012):3792−3803.
[Crossref] [Google Scholar] [PubMed]
- J.G. Luz, S. Antonysamy, S.L. Kuklish, B. Condon, M.R. Lee, et al. Crystal structures of mpges-1 inhibitor complexes form a basis for the rational design of potent analgesic and anti-inflammatory therapeutics, J Med Chem, 58(2015):4727−4737.
[Crossref] [Google Scholar] [PubMed]
- N. Uramaru, H. Shigematsu, A. Toda, R. Eyanagi, S. Kitamura, et al. Design, synthesis, and pharmacological activity of non allergenic pyrazolone-type antipyretic analgesics, J Med Chem, 53(2010):8727−8733.
[Crossref] [Google Scholar] [PubMed]
- J.O. Benjamin, G.M. Michael, Substrate-selective inhibition of cyclooxygeanse-2 by fenamic acid derivatives is dependent on peroxide tone, J Biol Chem, 291(2016):15069-15081.
[Crossref] [Google Scholar] [PubMed]
- M.T. Islam, P. Ray, A.B.R Khalipha, S. Hafiz Hassan, M.R. Khan, Molecular docking study of the phytol and its derivatives against COX-2 induced inflammation: A combined density functional study, Rec Res Sci Tech, 12(2020):1–5.
[Crossref] [Google Scholar] [PubMed]
- N.P. Pujan, P.K. Sivakumar, B. Kinjal, N.P. Chirag Saumya, K.P. Rakesh, et al. UM: Identification of promising compounds from curry tree with cyclooxygenase inhibitory potential using a combination of machine learning, molecular docking, dynamics simulations and binding free energy calculations, Mol Simln, 46(2020):812-822.
[Crossref] [Google Scholar] [PubMed]
- E. D. AlFadly, P.A. Elzahhar, A. Tramarin, S. Elkazaz, H. Shaltout, et al. RW: Tackling neuroinflammation and cholinergic deficit in Alzheimer's disease: Multi-target inhibitors of cholinesterases, cyclooxygenase-2 and 15-lipoxygenase, Eur J Med Chem, 167(2019):161-186.
[Crossref] [Google Scholar] [PubMed]
Copyright: © 2022 Neelaveni, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.