Research Letter - Journal of Drug and Alcohol Research ( 2022) Volume 11, Issue 1
Identification of Factors Regulating the Gene Expression of Alcohol Dehydrogenases in Hepatocellular Carcinoma
Jinghe Xie1, Yaqi Qiu2, Shuai Zhang3, Keqiang Ma4, Yimeng Ou5, Bailin Wang6, Weili Gu3, Honglin Chen2,7,8,9,10 and Yuyou Duan2,7,8,9,10*2Department of Medicine, South China University of Technology, China
3Department of Gastroenterology and Hepatology, South China University of Technology, China
4Department of Hepatobiliary Pancreatic Surgery, Huadu District People’s Hospital of Guangzhou, China
5Department of General Surgery, The First Affiliated Hospital of Guangdong Pharmaceutical University, China
6Department of General Surgery, Jinan University, China
7National Engineering Research Center for Tissue Restoration and Reconstruction, South China University of Technology, China
8Key Laboratory of Biomedical Engineering of Guangdong Province, South China University of Technology, China
9Key Laboratory of Biomedical Materials and Engineering of the Ministry of Education, South China University of Technology, China
10Innovation Center for Tissue Restoration and Reconstruction, South China University of Technology, China
Yuyou Duan, Innovation Center for Tissue Restoration and Reconstruction, South China University of Technology, China, Email: yuyouduan@scut.edu.cn
Received: 17-Dec-2021, Manuscript No. jdar-21-50028;;Accepted Date: Jan 07, 2022; Editor assigned: 20-Dec-2021, Pre QC No. jdar-21-50028 (PQ); Reviewed: 03-Jan-2022, QC No. jdar-21-50028; Revised: 07-Jan-2022, Manuscript No. jdar-21-50028 (R); Published: 14-Jan-2022, DOI: 10.4303/jdar/236157
Abstract
Background: Excessive alcohol consumption has been documented to increase the risk of liver hepatocellular carcinoma (HCC) development. Accordingly, a broad interest pointed to alcohol dehydrogenases (ADHs), which display essential roles in alcohol metabolism. Despite the relevance of ADHs expression and the prognosis of HCC has been estimated, so far, limited research concerning the factors that are responsible for the regulation of ADHs expression has been reported.
Methods: In this study, using The Cancer Genome Atlas (TCGA) and RegNetwork database, we predicted potential factors consisting of DNA methylation, gene copy number variations, and transcription factors (TFs) and microRNAs (miRNAs) that might impact ADHs gene expression in HCC.
Results: We found that DNA methylation induced the down regulated expression of ADH1B. Of note, our results implicated that gene copy number variation might not have effects on ADHs expression. Regarding TFs, we speculated that NFYA modulated ADH1C, E2F1 and TFAP2A regulated ADH6 expression based on their expression and prognostic value. Moreover, miR-185 and miR-561 might elicit the repression of ADH4, and miR-105 might impair ADH6 expression.
Conclusion: This study revealed that multiple factors, including DNA methylation, TFs and microRNAs, affect the expression of ADH family members, which provided new insights into discovering promising HCC-suppressive targets.
Introduction
Hepatocellular carcinoma (HCC), which frequently occurs in the patients with chronic liver inflammation, has been detected in approximately 90% of primary liver cancer [1]. HCC is well acknowledged as a lethal disease due to its high yearly mortality, leading to 830,000 deaths worldwide in 2020 [2]. Currently, the scientific community effort to develop a considerable number of diagnostic biomarkers has raised hope for timely diagnosis of HCC [3]. However, the majority of HCC patients are still detected in advanced stages, and thus are ineligible for receiving effective treatment [4]. Therefore, the request of elucidating the molecular mechanisms underpinning HCC onset and progression remains open.
Excessive alcohol consumption, one of the most common causes of liver cirrhosis, has been demonstrated to contribute to HCC development [5]. Previous study showed that genetic variation of alcohol dehydrogenases (ADHs) in advanced stage of alcoholic liver disease suppressed the activity of the enzymes, thus enhancing the amount of acetaldehyde [6]. Alcohol dehydrogenases (ADHs), consisting of class I (ADH1A, ADH1B and ADH1C), class II (ADH4), class III (ADH5), class IV (ADH6), and class V (ADH7), are a family of enzymes that play critical roles in alcohol metabolism by catalyzing the reversible oxidation of alcohol to acetaldehyde [7]. ADHs are encoded by seven ADH genes that locate in a segment of ∼370 kb on chromosome 4 (4q21) and are transcribed in the order of ADH7, ADH1C, ADH1B, ADH1A, ADH6, ADH4 and ADH5 [8, 9]. Of note, the gene expression of ADH1A, ADH1B, ADH1C, ADH4, and ADH6 has been identified to be strongly decreased in HCC patients, revealing the importance of ADH expression in association with the increased risk of liver cancer [10]. Nevertheless, the mechanisms responsible for the modulated level of ADH gene expression remains elusive.
In this study, we aimed to identify potential factors that exert profound influence on mediating ADH gene expression during HCC progression, which might provide novel therapeutic targets and prognostic factors for HCC. To this aim, we downloaded the expression, DNA methylation and copy number variation data from TCGA database and we analyzed the expression level of ADH members in tumor and normal samples. The correlation between DNA methylation and gene expression and the relationship between copy number variation and expression levels were performed as well. Also, we searched the TFs and miRNAs that regulated ADH members directly from RegNetwork database and depicted the effects to expression level of ADHs.
Materials and Methods
Acquisition of mRNA, transcription factors (TFs) and microRNAs (miRNAs) data
We downloaded liver hepatocellular carcinoma (HCC) mRNA, TFs and miRNA data from the TCGA database in December 2020, and obtained a total of 375 cancer tissues and 50 para-cancerous tissues. To collect expression data of TF and miRNA which regulated the ADH family members, we first used in house Perl scripts to merge FPKM matrix of all individuals, and then selected SRF, NFYA, HNF4A, CEBPB, E2F1, NFIC, SP1, TFAP2A, as well as hsa-mir-377, hsa-mir-181c, hsa-mir-185, hsa-mir-376c, hsa-mir-384, hsa-mir-410, hsa-mir-561, hsa-mir-657, hsamir- 944, hsa-mir-105, hsa-mir-299-3p, hsa-mir-498 and hsa-mir-522 from FPKM matrix by using Python script. The expression of hsa-mir-384 is zero, so it was not considered in the subsequent analysis. In addition, GSE28619 dataset from GEO, including 20 liver cancer tissues and 12 adjacent tissues, was used for external validation of ADHs expression. All of ADH members could be found in GSE28619 except ADH7.
DNA methylation of ADH genes in HCC
We downloaded the methylation data of 430 HCC samples from TCGA, and merged them into a matrix by using in house Perl script. The correlation analysis and Pearson correlated analysis were performed by using in house R script.
Copy number variation analysis
We downloaded gene level focal segment copy number of HCC samples form TCGA database, and selected the focal score of ADH family members by using in house R script. The boxplot of copy number variation and significance test was analyzed by using ggplot2 packages and Wilcox test respectively.
Prediction of TFs and miRNAs that regulate ADH family members
Potential TFs and miRNAs that regulate ADH family members were predicted based on the RegNetwork database (http://www.regnetworkweb.org/), which comprises the documented regulations among TFs, miRNAs and target genes from multiple databases and the potential regulations based on the TFs binding sites.
Identification of differentially expressed TFs and miRNAs
According to the TCGA nomenclature, the mRNA, TFs and miRNA expression data of HCC was divided into two groups including cancer tissues and adjacent non-tumor tissues. We used ggplot2 package in R version 4.0.2 for analyzing the difference between cancer tissues and adjacent non-tumor tissues. Then the Wilcox test was employed for significance test between tumor and non-tumor classes.
Prognostic values of TFs and miRNAs
We obtained vital status and overall survival (OS) information from the clinical data, and merged them with TFs and miRNAs expression data. We used survival package to perform non-parametric log-rank test on the high and low expression group of TFs and miRNAs. The significance of the P value was annotated on the Kaplan-Meier survival curve for OS. Multivariate Cox regression analysis was performed to evaluate the impact of TFs on survival time.
Statistical analysis
All statistical analyses were processed with R 4.0.2 software. The compare between two groups was performed by using Wilcox test. P<0.05 was considered as statistically significant. Pearson correlation coefficient was used to measure the degree of correlation between DNA methylation and expression level.
Results
ADH family members were down-regulated in human HCC
Previous study assessed the gene expression of ADH family members in TCGA data, and demonstrated that ADH1A, ADH1B, ADH1C, ADH4, and ADH6 expression were significantly decreased in HCC tissues compared with normal controls [10]. In the present study, we further analyzed the transcriptome expression level of all ADH genes in 20 liver cancer and 12 tumor adjacent normal tissues by employing GEO dataset. Consistently, we observed a remarkable reduction in ADH1A, ADH1B, ADH4, and ADH6 expression in HCC as compared to normal tissues (Figure 1). Of note, there were no statistically significant differences in the expression of ADH1C and ADH5 between the two groups (Figure 1A).
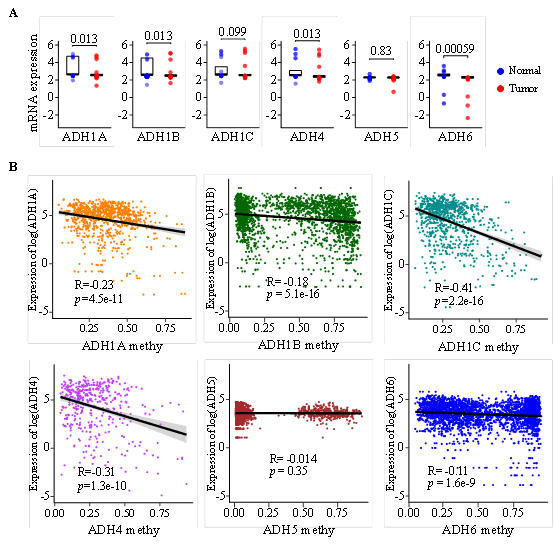
Figure 1: (A) ADH family members were down-regulated in human HCC. mRNA expression of six ADH members in normal tissues (n=12) and HCC tissues (n=20) derived from GEO database. (B) The correlation analysis between gene expression and DNA methylation derived from TCGA data. The X axis presented DNA methylation level of ADH1A, ADH1B, ADH1C, ADH4, ADH5 and ADH6, and the Y axis presented gene expression level that quantified as log (FPKM).
Correlation between ADHs gene expression and methylation
In attempt to elucidate the mechanism underlying the modulated expression of different ADH genes in HCC, we first evaluated DNA methylation level of all ADH genes from 41 normal and 374 HCC in TCGA. As shown in Figure 2, the methylation level of ADH1A, ADH1B, ADH1C, ADH4 and ADH6 were negatively correlated with their gene expression. Moreover, no significant association between ADH5 expression and methylation was detected (Figure 1B). It has been illustrated that multiple methylated CpG sites located in promoter region could induce transcriptional silencing of genes [11]. We found that cg24368912 within the sequence of ADH1B promoter, and cg09112717, cg13256891, cg15443145, cg21548116 and cg25073725 in ADH5 promoter had significantly higher methylated levels in HCC than normal controls (Figure 2). Conversely, DNA methylation levels at cg03806087, cg23949936 in ADH1A promoter, cg00689360 in ADH1C promoter, cg12011299 in ADH4 promoter, and cg00268009 in ADH5 promoter were evidently reduced in HCC as compared with healthy samples (Figure 2).
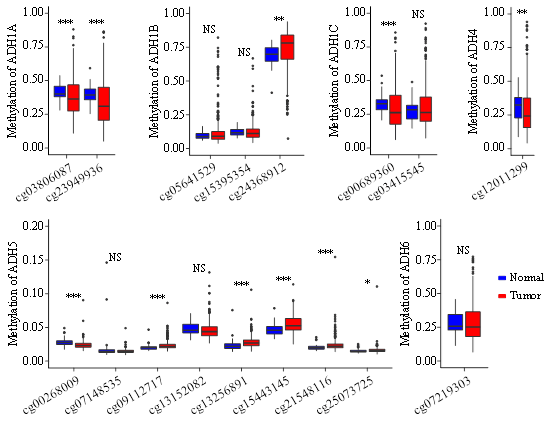
Figure 2: The methylation levels of CpG sites within the promoter region of ADH genes between normal hepatocytes and HCC samples.
Correlation between ADHs gene expression and gene copy number
Being one of the predominant forms of genetic alterations, copy number variation has been documented to play a vital role in regulating gene expression during tumor progression [12]. Accordingly, we further performed an analysis of the gene level focal segment copy number derived from 190 HCC cases in TCGA data. However, we did not observe significant alterations of gene copy number for the gene expression of ADH1A, ADH1B, ADH1C, ADH4, and ADH6 in HCC (Figure 3). In contrast, ADH5 expression was markedly downregulated with a reduced gene copy number (Figure 3). Thus, our data implicated that copy number variation might not apparently affect the gene expression level of ADH genes in HCC subjects.
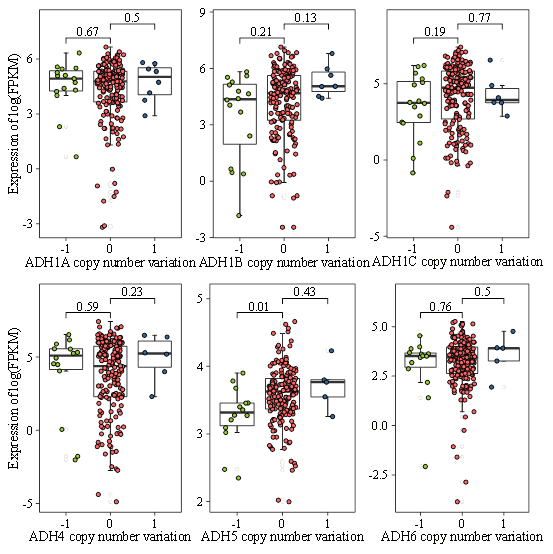
Figure 3: The correlation analysis between ADH gene expression and gene copy number derived from TCGA data. The X axis presented gene copy number of ADH1A, ADH1B, ADH1C, ADH4, ADH5 and ADH6, and the Y axis presented gene expression level quantified as log (FPKM).
Identification of the potential TFs that regulate ADH gene expression
To better clarify other regulatory factors for ADH gene expression, we conducted the prediction of potential TFs, which are proteins that participate in transcriptional regulation of genes through binding specific DNA sequences [13]. From RegNetwork database, SRF, NFYA and HNF4A were identified as TFs that target ADH1B, ADH1C and ADH4 respectively (Figure 4A). Interestingly, five TFs (TFAP2A, CEBPB, E2F1, SP1 and NFIC) were all recognized as regulators of ADH6 gene expression (Figure 5A). Next, we ascertained the gene expression levels of these predictive TFs in HCC tissues in contrast with normal tissues. Remarkably, all the predictive TFs were up regulated in HCC except CEBPB (Figure 4B). Furthermore, we examined the correlation between above TFs and overall survival. The results of Kaplan-Meier analysis revealed that HCC patients with high expression levels of NFYA, E2F1 and TFAP2A were strongly associated with shortened overall survival (Figure 5).
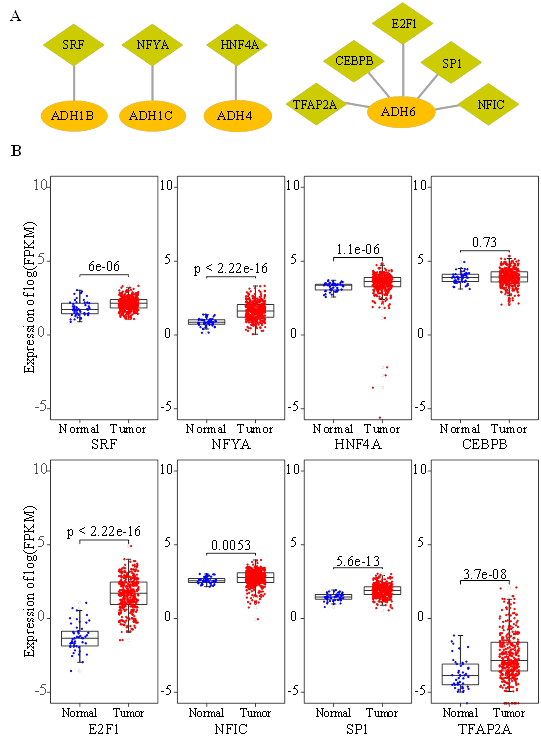
Figure 4: Identification of the potential TFs that regulate ADH gene expression. (A) Potential TFs that regulate ADH family members were predicted by utilizing RegNetwork database. (B) The gene expression levels of TFs in normal hepatocytes and HCC samples derived from TCGA data.
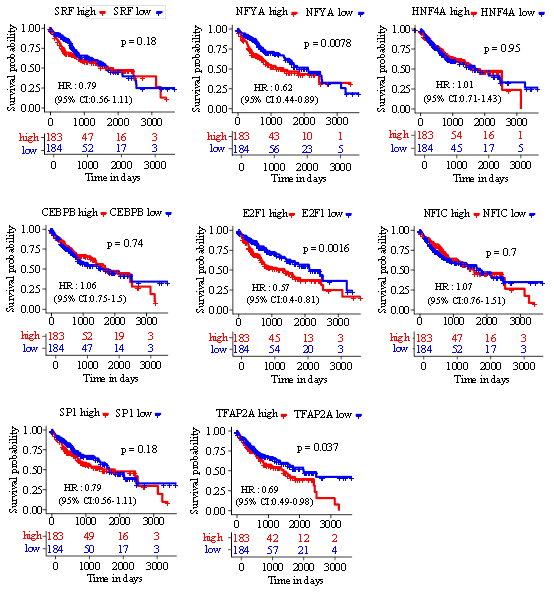
Figure 5: Kaplan-Meier overall survival curve was shown according to high and low expression of TFs in HCC patients derived from TCGA data. (log-rank<0.05).
Identification of the potential miRNAs that regulate ADH gene expression
As small non-coding RNA molecules, miRNAs post-transcriptionally modulate gene expression [14]. Thus, we also predicted the potential miRNAs that were involved in ADH gene regulation by employing RegNetwork database. miR- 377 and miR-944 were discovered to respectively impact ADH1B and ADH5 expression (Figure 6A). Notably, we found that ADH4 was mediated by 7 miRNAs, including miR-181c, miR-185, miR-376c, miR-410, miR-561, miR- 657 and miR384 (Figure 6A). In addition, ADH6 was observed to be regulated by miR-105, miR-299-3p, miR-498 and miR-522 (Figure 6A). To understand the effects of the predictive miRNAs exerted on HCC progression, we initially measured their expression levels. We noticed that the expression of miR-377, miR-376c, miR-410, miR-299-3p were evidently suppressed in HCC subjects in comparison with normal controls (Figure 6B). On the contrary, miR- 185 and miR-105 expression were significantly enhanced in HCC (Figure 6B). Further analysis of the association between miRNAs expression and survival time indicated that high expression of miR-561 and miR-498 were both predictors of poor overall survival in HCC patients (Figure 7).
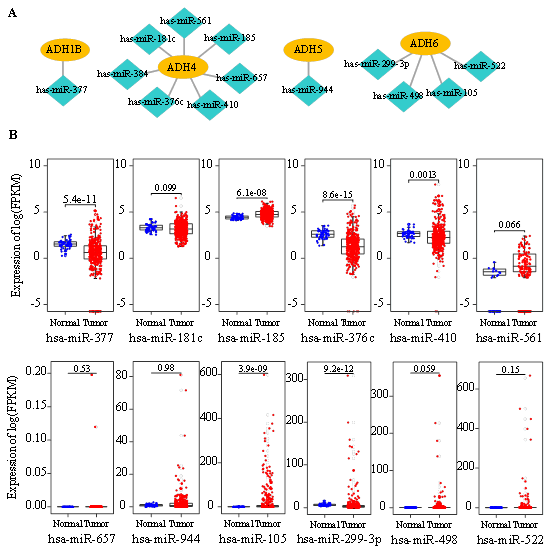
Figure 6: Identification of the potential miRNAs that regulate ADH gene expression. (A) Potential miRNAs that regulate ADH family members were predicted by utilizing the RegNetwork database. (B) The expression levels of miRNAs in normal hepatocytes and HCC samples derived from TCGA data.
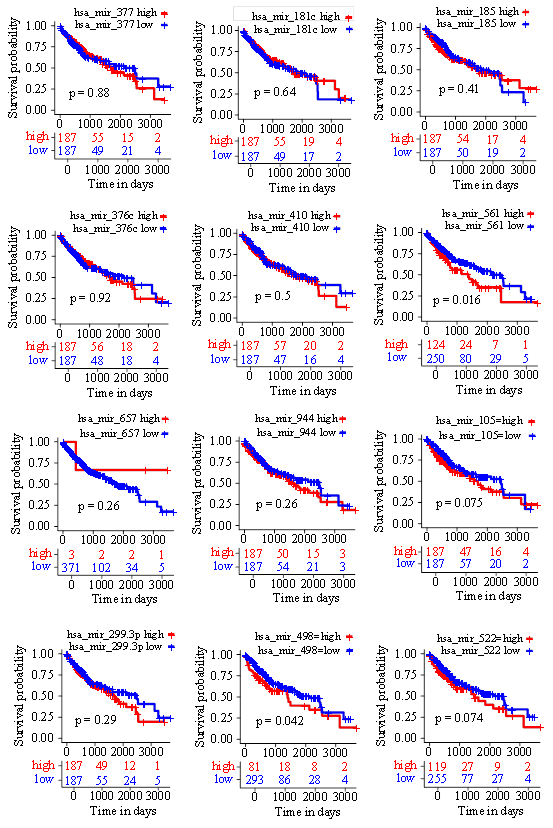
Figure 7: Kaplan-Meier overall survival curve was shown according to high and low expression of miRNAs in HCC patients derived from TCGA data. (log-rank<0.05).
Discussion
The evidence that ADH1A-ADH6 are widely expressed across human liver and related to liver cancer risk raised interest in investigating the prognostic value of these genes [10,15]. Recent results have implicated that ADH1A, ADH1C, ADH4 and ADH6 were positively correlated with the prognosis of HCC patients, suggesting that these ADH members might serve as novel diagnostic and prognostic markers for HCC [10,16]. Although the roles of ADH genes involved in liver carcinogenesis have been explored, little is known about the mechanisms underlying the modulated level of ADHs expression. Herein, for the first time, we identified potential factors that affect ADHs expression during HCC progression.
DNA methylation has been well characterized as a critical regulatory mechanism for gene expression [17]. Indeed, our data revealed that the gene expression of ADHs without ADH5 were all negatively associated with methylation level. It has been demonstrated that upstream of ADH1A, ADH1B and ADH1C are methylated in the HCC cell line HepG2 [18]. Consistently, we found that CpG island in the promoter region of ADH1B had markedly higher methylation level in HCC than normal hepatocytes samples, indicating that DNA methylation might be responsible for the down-regulated expression of ADH1B. However, for the repressed expression of ADH1A, ADH1C, ADH4 and ADH6 in HCC, DNA methylation might not be the major mechanism.
In addition, we also examined the effects of gene copy number variations, which include excess or deficiency of sections of DNA sequence, on ADHs gene expression [19]. In our study, we demonstrated that only ADH5 gene copy number variations (deletions) were more frequent in HCC patients. A recent study reported that no significant difference was detected for gene copy number variations of ADH1A, ADH1B and ADH1C between alcohol related liver cirrhosis patients and healthy samples [20]. In line with the above findings, our results indicated that no statistical significance in gene copy number variations of ADH1A, ADH1B, ADH1C, ADH4 and ADH6 in HCC, suggesting that gene copy number variations might not profoundly influence ADHs expression.
TFs and miRNAs, as critical trans-regulators of gene expression, have been documented to function in the transcriptional and post-transcriptional levels, respectively [21]. In the context of carcinogenesis, TFs and miRNAs might exert oncogenic or tumor-suppressive role in modulating tumor progression [22]. By assessing the expression and prognostic values of predicted TFs, we speculated that NFYA, E2F1 and TFAP2A might act as regulators for ADH1C and ADH6 expression respectively. Regarding miRNAs, miR-185 and miR-561 might suppress ADH4 expression, and miR-105 might inhibit ADH6 expression. Further research is necessary to verify these findings.
To the best of our knowledge, our study for the first time identified the factors that mediate ADHs gene expression in the pathogenesis of HCC, which will be valuable in more understanding the mechanisms underpinning ADHs expression and could be applied to the prediction of HCC-suppressive targets.
Conclusion
We found that ADH family members were infected by disparate factors. ADH1B was down regulated by DNA methylation, while ADH1C and ADH4 was elicited by TFs and miRNA respectively. The expression of ADH6 was modulated by TFs and miRNA at the same time.
Acknowledgements
Not applicable.
Authors Contributions
YD and HC supervised the study. JX and SZ collected data and conducted the expression analysis, prognosis analysis and statistical analysis. YQ and KM performed the miRNA and TF interaction analysis and methylation analysis. HC, YQ and YO wrote the paper together. BW and WL critically reviewed and revised the manuscript. All authors read, edited, and approved the final paper.
Funding
This work was supported in part by Research Starting Funding of South China University of Technology (D6181910, D6201880, K5180910, and K5204120 to Y.D.), by Research Agreement between South China University of Technology and Guangzhou First People’s Hospital (D9194290 to Y.D.; PT31900976 to H.C.), by the National Natural Science Foundation of China (No. 31900976 and No. 32071360, to H.C.).
Availability of Data and Material
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
All authors have reviewed the manuscript and agree to publish it in its current form.
Competing Interests
The authors declare that they have no competing interests.
Conflict Of Interest Statement
The authors declare that they have no competing interests.
References
- M. F. Chedid, C. R. P. Kruel, M. A. Pinto, T. J. M. Grezzana-Filho, I. Leipnitzet al., Hepatocellular Carcinoma: Diagnosis and Operative Management, Arq Bras Cir Dig, 30 (2017), 272-278.
- J. Ferlay, M. Colombet, I. Soerjomataram, D. M. Parkin, M. Pineroset al., Cancer statistics for the year 2020: An overview, Int J Cancer, 149 (2021), 778-789.
- K. Singh, R. Kumar,A. K. Pandey, Hepatocellular Carcinoma: Causes, Mechanism of Progression and Biomarkers, Curr Chem Genom Transl Med, 12 (2018), 9-26.
- S. Colagrande, A. L. Inghilesi, S. Aburas, G. G. Taliani, C. Nardiet al., Challenges of advanced hepatocellular carcinoma, World J Gastroenterol, 22 (2016), 7645-7659.
- H. McKillop, L. W. Schrum,K. J. Thompson, Role of alcohol in the development and progression of hepatocellular carcinoma, Hepat Oncol, 3 (2016), 29-43.
- Hyun, J. Han, C. Lee, M. Yoon,Y. Jung, Pathophysiological Aspects of Alcohol Metabolism in the Liver, Int J Mol Sci, 22 (2021).
- H. J. Edenberg, The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants, Alcohol Res Health, 30 (2007), 5-13.
- D. Li, H. Zhao,J. Gelernter, Further clarification of the contribution of the ADH1C gene to vulnerability of alcoholism and selected liver diseases, Hum Gen, 131 (2012), 1361-1374.
- S. Jairam,H. J. Edenberg, Single-nucleotide polymorphisms interact to affect ADH7 transcription, Alc clin and exp res, 38 (2014), 921-929.
- X. Liu, T. Li, D. Kong, H. You, F. Konget al., Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma, BMC Cancer, 20 (2020), 1204.
- M. Esteller, CpG island hypermethylation and tumor suppressor genes: A booming present, a brighter future, Oncogene, 21 (2002), 5427-5440.
- Bhattacharya, R. D. Bense, C. G. Urzua-Traslavina, E. G. E. de Vries, Matm van Vugtet al., Transcriptional effects of copy number alterations in a large set of human cancers, Nat Commun, 11 (2020), 715.
- S. R. Grossman, J. Engreitz, J. P. Ray, T. H. Nguyen, N. Hacohenet al., Positional specificity of different transcription factor classes within enhancers, Proc Natl Acad Sci USA, 115 (2018), E7222-E7230.
- O'Brien, H. Hayder, Y. Zayed,C. Peng, Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation, Frn Endcrl (Lausanne), 9 (2018), 402.
- H. J. Edenberg,J. N. McClintick, Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review, Alcoholism, Clin and exp res, 42 (2018), 2281-2297.
- R. R. Wei, M. Y. Zhang, H. L. Rao, H. Y. Pu, H. Z. Zhanget al., Identification of ADH4 as a novel and potential prognostic marker in hepatocellular carcinoma, Med Oncol, 29 (2012), 2737-2743.
- J. L. Miller,P. A. Grant, The role of DNA methylation and histone modifications in transcriptional regulation in humans, Subcell Biochem, 61 (2013), 289-317.
- O. Dannenberg, H. J. Chen, H. Tian,H. J. Edenberg, Differential regulation of the alcohol dehydrogenase 1B (ADH1B) and ADH1C genes by DNA methylation and histone deacetylation, Alcsm clin exp res, 30 (2006), 928-937.
- J. Sebat, B. Lakshmi, J. Troge, J. Alexander, J. Younget al., Large-scale copy number polymorphism in the human genome, Sci, 305 (2004), 525-528.
- P. Ayuso, E. Garcia-Martin, J. A. Cornejo-Garcia, J. A. G. Agundez,J. M. Ladero, Genetic Variants of Alcohol Metabolizing Enzymes and Alcohol-Related Liver Cirrhosis Risk, J Pers Med, 11 (2021).
- G. Qin, S. Mallik, R. Mitra, A. Li, P. Jiaet al., MicroRNA and transcription factor co-regulatory networks and subtype classification of seminoma and non-seminoma in testicular germ cell tumors, Sci Rep, 10 (2020), 852.
- Zhao, P. Kim, R. Mitra, J. Zhao,Z. Zhao, TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes, Nucl Acid Res, 44 (2016), D1023-1031.