Research - Journal of Drug and Alcohol Research ( 2023) Volume 12, Issue 1
In Silico based Identification of Novel Phytomolecules from Piper Longum for Drug Discovery against Cox-2
Indrajeet Singh1, Hara Prasad Mishra2, Pravesh Kumar1 and Ajay Kumar1*2Department of Medical Sciences, University of Delhi, Delhi, India
Ajay Kumar, Department of Biotechnology, Rama University, Mandhana, Kanpur, Uttar Pradesh, India, Email: ajaymtech@gmail.com
Received: 31-Jan-2023, Manuscript No. JDAR-23-91987; Editor assigned: 02-Feb-2023, Pre QC No. JDAR-23-91987 (PQ); Reviewed: 16-Feb-2023, QC No. JDAR-23-91987; Revised: 21-Feb-2023, Manuscript No. JDAR-23-91987 (R); Published: 28-Feb-2023, DOI: 10.4303/JDAR/236223
Abstract
Background: Acquired drug resistance is one of the key clinical problems and the main downsides of chemotherapy regimens for colorectal cancer. Using blend regimens, which mix conventional chemotherapeutic medications through plant-derived natural compounds derived from food or vegetation, is one of the most popular and efficient ways to overcome drug resistance and re-sensitize chemotherapy-resistant cells. Colorectal cancer is amongst the most common cancers. Despite improvements in its therapies, a fuller understanding of the molecular processes and genetic contribution to colorectal cancer will play a crucial role in the development of innovative and targeted treatments with improved safety profiles. In this review, the association between piperlongumine and Cyclooxygenases- 2 colorectal carcinogenesis, as well as potential mechanisms of action, are investigated. Cyclooxygenases-2 participates in colorectal cancer and other malignancies have drawn a lot of interest because of their crucial role in decreasing, preventing, or altering the itinerary of the illness. The greater selectivity on certain macromolecular targets and significant advancements in colorectal cancer therapies with in-silico research were also presented. Additionally, by using in-silico approaches for target identification and medication repurposing, as well as molecular de novo synthesis, selective multi-target agents have been made simpler. The molecular docking investigation of piperlongumine with various apoptotic proteins implicated in colorectal cancer is documented. The data demonstrate that piperlongumine has ideal binding properties with Cox-2 for further investigation in this context.
Methodology: In current study we considered medicinal plants and their phytochemicals to reveal their interaction pattern with crucial enzymes Cox-2 that play an important role in cancer. In current study we aimed to conduct in-silico analysis for revealing the anti-inflamaation nature of phytochemicals piperlongumine. This study also involves molecular docking, Pharmacophore model, Lipinski drug analysis, and fundamental ADME analysis of phytochemicals that assisted us in developing core medicinal background of phytochemicals.
Results: The molecular docking investigation of piperlongumine with various apoptotic proteins implicated in colorectal cancer is documented. The data demonstrate that piperlongumine has ideal binding properties with Cox-2 for further investigation in this context.
Conclusion: This study advocates the use of substances containing piperlongumine as possible colorectal cancer therapies. This lead chemical will be useful to the scientific community and might help in the search for novel colorectal cancer therapies.
Keywords
Molecular docking; Piperlongumine; COX-2; Molecular dynamics simulation; Interaction analysis; Natural product
Abbreviations
(Cox-2) Cyclooxygenases-2; (PL) Piperlongumine; (BCL2) B-cell leukemia/lymphoma 2; (ROS) Reactive Oxygen Species; (MAPK) Mitogen-activated Protein Kinase; (GSTP1) Glutathione S-transferase P1; (NSAIDs) Nonsteroidal Anti-Inflammatory Drugs; (TxA2) Thromboxane A2; (RMSD) Root Mean Square Deviation; (SMAD4) Mothers against Decapentaplegic Homolog 4; (CRC) Colorectal Cancer; (ADME) Absorption, Distribution, Metabolism, Excretion and Toxicity; (NMR) Nuclear Magnetic Resonance
Introduction
Gastrointestinal cancer (colorectal cancer) is the third most typical malignancy and the third leading cause of cancer- related death in the United States [1]. The prevention and treatment of colorectal cancer are extremely important for the public’s health because of the disease’s relatively high incidence and resistance to currently available anti-cancer medicines. Reactive oxygen species (ROS) have been shown to be crucial in both carcinogenesis and apoptosis, despite the fact that they are two distinct events. While medications, harmful substances, and UV radiation may all cause ROS, intracellular ROS is formed as a by-product of regular metabolism. Different cellular activities are triggered by variations in intracellular ROS levels. ROS functions as a signalling molecule for numerous signal transductions at low doses [2,3]. But at large quantities it is fatal [4]. The cells engage their antioxidant machinery, which is made up of antioxidant enzymes and antioxidant metabolites, to combat the intracellular oxidative stress. Cancer cells typically contain high levels of antioxidant system and increased ROS [5]. Since too much oxidative stress causes apoptosis, which leads to cell death, Excessive ROS generation has been linked to the anticancer activity of medications like paclitaxel, 2-methoxyestradiol and trisenox [6-8]. Observational studies that regularly demonstrate a 40%-50% reduction in CRC prevalence among long term users of non-steroidal anti-inflammatory medicines (NSAIDs) have raised interest in cyclooxygenase (COX) as a therapeutic target for CRC [9-11]. The NSAID’s most well-defined molecular target, COX, controls how pro-inflammatory cytokines (prostaglandins) and related eicosanoids are made from arachidonic acid. The enzymes constitutively expressed COX-1 and induced COX-2 are examples of COX isoforms. The prostaglandin Endoperoxidase synthase COX-2, also known as prostaglandin H synthase-1 or COX-2 is activated in response to cytokine production, growth hormones, and tumour regulators. In contrast to healthy epithelial cells, COX-2, but not COX-1, has also been demonstrated to be overexpressed in an estimated 40% of human colorectal adenomas as well as 80% of adenocarcinomas [12]. COX-2 production has been seen in gastrointestinal adenomas in mice treated with azoxymethane (AOM) in colorectal cancer animal models [3] as well as the ApcMin (1) mice, which contains a completely penetrant, dominant mutation in the murine APC gene [4]. COX enzymes participate a significant role in catalysing the synthesis of PGs from arachidonic acid [12-15]. Arachidonic acid is changed by COX from membrane phospholipids into PGG2 and subsequently PGH2 after being released. The action of certain PG synthases then converts PGH2 into one of numerous prostaglandins with similar structural properties, such as PGE2, PGD2, PGF2, PGI2, and thromboxane A2 (TxA2). A “housekeeping” gene, the constitutive COX-1 isoform contributes to the preservation of tissue homeostasis. It is generated in the majority of healthy tissues, including the gastrointestinal tract’s mucosa, where it is thought to guard against gastric injury. In the late 1980’s, researchers discovered and cloned COX-2, a second COX enzyme. Cells altered with the v-src oncogene [16] or exposed to phorbol esters [17] exhibited COX-2 expression. Growth factors, proinflammatory cytokines, and tumour promoters can all stimulate COX-2, an intermediate response gene that produces a cytoplasmic protein with a molecular weight of 71 kDa [18]. COX-2 is over expressed in human colorectal neoplasms as well as in a number of other epithelial malignancies [1,19] and is up-regulated in areas of inflammation [20]. Both COX-1 and COX-2 isoform activity is inhibited by conventional NSAIDs. It is thought that COX-1 and prostaglandin production suppression is the cause of NSAID-induced gastric mucosal damage, renal toxicity, and antiplatelet effects [21,22]. The anti-inflammatory and analgesic effects of NSAIDs are linked to their capacity to inhibit COX-2 since it is activated at sites of inflammation. As a result, there are two types of COX isoforms: constitutive COX-1, which is engaged in maintaining normal tissue homeostasis, and inducible COX-2, which is increased in inflammatory areas and in colorectal tumours. While COX-2 inhibitors are specific for the COX-2 enzyme, NSAIDs inhibit both COX isoforms. To precisely inhibit the COX-2 isoform, Sinicrope and Gill created a new family of NSAIDs called coxibs.
About 80% of human cancers and 40% of colorectal adenomas over manifested COX-2 proteins in comparison to normal mucosa [1,22]. Although some stromal staining is seen, COX-2 localises mostly to the cytoplasm of malignant epithelial cells in immunohistochemistry. The COX-2 status of normal epithelium is, however, negative. Concerning the prevalence of COX-2 protein appearance in human colorectal tumours, results have been similar [22]. Furthermore, it has been shown that colorectal tumours express more COX-2 messenger RNA (mRNA) than does healthy mucosa [1]. However, COX-1 levels are comparable in the healthy mucosa and the colorectal tumour, which is consistent with its constitutive expression. Although to a lower extent than in epithelial cells, stromal cell expression of COX-2 in macrophages, fibroblasts, and endothelial cells has been seen in these malignancies. The malignant epithelial cells are also the main source of COX-2 over expression in other epithelial malignancies, such as those of the breast pancreatic oesophagus stomach and lung.
A lipid-soluble alkaloid amide known as piperlongumine (PL) was discovered in the long pepper (Piper longum L.). Piper longum is a fragrant woody root. Numerous alkaloids with potent anti-inflammatory, anti-angiogenic, anti-lipidemic, and antifungal properties are present in piper longum extract. A common substance, PL, was revealed by Raj et al. to destroy cancer cells of many sources in a very precise way. According to a research, PL selectively induces cancer cell death. According to research on the route of action, PL may prevent glutathione S-transferase P1 (GSTP1) from acting as an anticancer agent, particularly through producing reactive oxygen species (ROS). Contrary to several other drugs that produce ROS, the researchers claim that PL causes apoptotic cell death. The purpose of this study was to determine how the proteins PL and Cox-2 interacted with colon cancer and to determine whether PL might modify Cox-2-mediated apoptosis.
Methods and Materials
Receptor selection and pocket prediction
The Cyclooxygenase-2 (Prostaglandin G/H synthase-2) (Uniprot ID: P35354) has 604 amino acids in length. As crystallographic structure has many missing side chains and bond redundancy, we select a 3D structure constructed from the AlphaFold AI system for docking analysis. The AlpahFold model aligned with available experimental structures which showed zero RMSD. This suggests the AlphaFold model has exact folding of the secondary structure except for terminal regions (N-terminal: 1-18 and C-terminal: 565-604). The active site region was predicted using the CastP web server the selected active region ion has an 893.648A2 area containing the following residues; Tyr148, Ala199, Gln203, His207, Phe210, Lys211, Thr212, His214, Asn382, Tyr385, His386, Leu391, and Phe395. The COX-2 structure was minimized using the YASARA energy minimization server that uses the YASARA force field with Monte Carlo minimization.
Toxicity prediction and preparation
The piperlongumine was downloaded from the PubChem database and selected for docking calculation against COX- 2. The piperlongumine 3D structure was prepared by adding hydrogen atoms and AM1-BCC-based charges computed using the ANTEHAMBER module. The prepared structure is minimized by applying 100 steps of steepest descent followed by 10 steps of conjugate gradient algorithm. The preparation and minimization was performed by Chimera v1.12. ProTox-II, machine-learning based program used to determine the physicochemical properties of piperlongumine (https://tox-new.charite.de/protox_II/index. php?site=home) shown in Table 1. The ADMET and toxicity of piperlongumine was analysed using ADMET Lab (https://admet.scbdd.com/home/index/) shown in Table 2 as well as toxicity profile based on predicted values of piperlongumine shown in Table 3.
Table 1: Physicochemical properties of piperlongumine collected from PubChem, ADMETLab, and ProTox-II program.
| S. No. | Name | Piperlongumine |
|---|---|---|
| 1. | Mol Weight | 317.34 |
| 2. | Number of hydrogen bond acceptors | 25 |
| 3. | Number of hydrogen bond donors | 0 |
| 4. | Number of atoms | 42 |
| 5. | Number of bonds | 43 |
| 6. | Number of rotatable bonds | 6 |
| 7. | Molecular refractivity | 89.47 |
| 8. | Topological Polar Surface Area | 65.07 |
| 9. | octanol/water partition coefficient(logP) | 1.98 |
| 10. | LogS (Solubility) | -3.002 |
| 11. | LogD7.4 (Distribution Coefficient D) | 0.739 |
| 12. | Predicted LD50 | 1180 mg/kg |
| 13. | Predicted Toxicity Class | 4 out of 6 |
| 14. | Average similarity | 75.84% |
| 15. | Prediction accuracy | 69.26% |
Table 2: ADMET properties prediction using ADMETLab.
| Classification | Target | Prediction | Probability |
|---|---|---|---|
| Absorption | Papp (Caco-2 Permeability) | -4.432 cm/s | - |
| Pgp-inhibitor | + | 0.513 | |
| Pgp-substrate | - - - | 0.279 | |
| HIA (Human Intestinal Absorption) | + | 0.62 | |
| F (20% Bioavailability) | + | 0.525 | |
| F (30% Bioavailability) | + | 0.619 | |
| Distribution | PPB (Plasma Protein Binding) | 76.88% | - |
| VD (Volume Distribution) | -0.324 L/kg | - | |
| BBB (Blood–Brain Barrier) | +++ | 0.915 | |
| Metabolism | P450 CYP1A2 inhibitor | +++ | 0.968 |
| P450 CYP1A2 Substrate | + | 0.545 | |
| P450 CYP3A4 inhibitor | - - - | 0.069 | |
| P450 CYP3A4 substrate | + | 0.506 | |
| P450 CYP2C9 inhibitor | - - - | 0.048 | |
| P450 CYP2C9 substrate | - | 0.394 | |
| P450 CYP2C19 inhibitor | - - - | 0.109 | |
| P450 CYP2C19 substrate | - | 0.484 | |
| P450 CYP2D6 inhibitor | - - - | 0.25 | |
| P450 CYP2D6 substrate | + | 0.511 | |
| Elimination | T1/2(Half Life Time) | 1.412 h | >0.5 h |
| CL (Clearance Rate) | 1.571 mL/min/kg | - | |
| Toxicity | hERG (hERG Blockers) | + | 0.624 |
| H-HT (Human Hepatotoxicity) | ++ | 0.826 | |
| AMES (Ames Mutagenicity) | - | 0.304 | |
| SkinSen (Skin sensitization) | - - - | 0.24 | |
| LD50 (LD50 of acute toxicity) | 2.31 -log mol/kg (1554.269 mg/kg) | - | |
| DILI (Drug Induced Liver Injury) | - | 0.382 | |
| FDAMDD (Maximum Recommended Daily Dose) | + | 0.548 |
Table 3: Toxicity profile based on predicted values of Piperlongumine using ProTox-II, Inactive means no toxicity found while Active means there is some toxicity available based on predicted data.
| Classification | Target | Prediction | Probability |
|---|---|---|---|
| Organ toxicity | Hepatotoxicity | Inactive | 0.79 |
| Toxicity endpoints | Carcinogenicity | Inactive | 0.52 |
| Immunotoxicity | Active | 0.99 | |
| Mutagenicity | Inactive | 0.69 | |
| Cytotoxicity | Inactive | 0.62 | |
| Tox21-Nuclear receptor signalling pathways | Aryl hydrocarbon Receptor | Inactive | 0.88 |
| Androgen Receptor | Inactive | 0.94 | |
| Androgen Receptor Ligand Binding Domain | Inactive | 0.97 | |
| Aromatase | Inactive | 0.91 | |
| Estrogen Receptor Alpha | Inactive | 0.89 | |
| Estrogen Receptor Ligand Binding Domain | Inactive | 0.95 | |
| PPAR-Gamma | Inactive | 0.96 | |
| Tox21-Stress response pathways | nrf2/ARE | Inactive | 0.93 |
| Response component for the heat shock factor | Inactive | 0.93 | |
| Mitochondrial Membrane Potential | Inactive | 0.85 | |
| Phosphoprotein (Tumor Suppressor) p53 | Inactive | 0.9 | |
| ATPase family | Inactive | 0.94 |
Pharmacophore model
Using the PharmMapper online web server, the piperlongumine pharmacophore features are predicted. According to their normalised fit score with expected attributes, the 300 protein targets were ranked.
Molecular docking
The Piperlongumine pose prediction against COX-2 receptor structure was subjected to a molecular docking study performed by Autodock v4.4. The COX-2 structure was prepared by adding polar hydrogen atoms, charges computed by Gastegier followed by the added Kollman charge along with AD4 atoms. The piperlongumine structure was prepared by defining the root and number of torsion angles. The autodock format is used to preserve both prepared structures (PDBQT). The expected binding location was the centre of the grid, which included grid boxes of 60×60×60. A genetic search method was used to produce the docked file, and the Lamarckian genetic algorithm saved the outcome. Based on the lowest free binding energy from each cluster, the top position was chosen. The ChimeraX visualizer conducted the investigation of protein-ligand interactions.
Molecular dynamics simulation
A 100 ns molecular dynamics simulation of the COX-2 in Apo form and in complex with Piperlongumine was run on Gromacs v2020. The ACPYPE software created the Piperlongumine topology. Both systems (COX-2 apo and COX-2 complex) were created by introducing hydrogen atoms and charges while using the explicit TIP3P water solvent and Charmm36 force field. Both systems utilised the triclinic box periodic box conditions. For both system minimizations, the 5000-step steepest descent and conjugate gradient algorithms were employed. NPT and NVT ensemble class were used to equilibrate the system at 300 K for 100 ps.
Free energy analysis
The Piperlongumine complex with COX-2 binding free energy was analyzed using gmx_MMPBSA v1.4.3 scripts. Total 500 frames were selected from simulation trajectories by removing PBC conditions to compute the binding free energies.
Results
Toxicity prediction
The Piperlongumine toxicity was predicted using 17 toxicity models available in the ProTox-II program.
Pharmacophore model
The PharmMapper showed more than 4 pharmacophore features calculated from ~300 protein targets accessible in the Protein data bank. The good score lies at 2.862 while the normalized fit score lies at 0.954. These values indicated that piperlongumine has more hydrophobic features compared to others such as Aromatic, Number of positive and negative features. The presence of a hydrophobicity region indicates that piperlongumine can bind where the tunnel-like region is present in the protein structure shown in Figure 1.
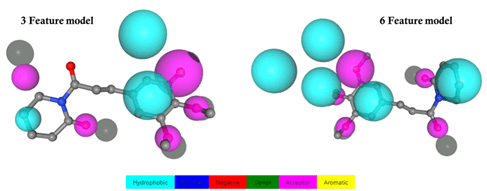
Figure 1: The predicted pharmacophore feature model of piperlongumine. The hydrophobic feature is present in more numbers >3 while the hydrogen bond acceptor is less than <2 in both 3 and 6 feature models.
Molecular docking analysis of selected proteins
The piperlongumine binding affinity was checked by docking calculation against selected 13 cancerous proteins of humans (Table 4). The 13 proteins were prepared and the binding region was selected based on the CastP server. The grid and box size were adjusted based on the predicted binding region (Table 5). The docking calculation showed that COX-2, JAK1, JAK2, and MCL-1 proteins have the lowest binding free energy compared to other proteins.
Table 4: Active site details selected for molecular docking.
| Protein name | PDB ID | Grid Spacing | Grid size | Grid centre | ||||
|---|---|---|---|---|---|---|---|---|
| X | Y | Z | X | Y | Z | |||
| BAX | 4S0O | 1 | 16 | 24 | 32 | 2.57 | 7.04 | 15.44 |
| COX-2 | 1CX2 | 1 | 21 | 25 | 25 | 28.68 | 28.6 | 9.29 |
| caspase-9 | 2AR9 | 1 | 22 | 22 | 20 | 15.73 | 40.9 | 0.99 |
| β-catenine | 1G3J | 1 | 25 | 27 | 21 | 13.97 | 74.21 | 61.56 |
| JAK1 | 4EI4 | 1 | 24 | 25 | 25 | 6.37 | 54.23 | 9.63 |
| JAK2 | 3KRR | 1 | 18 | 21 | 25 | 17.57 | 20.06 | -3.04 |
| STAT3 | 4ZIA | 1 | 25 | 20 | 25 | 138.9 | 61.12 | 126.79 |
| GSK3 | 5HLP | 1 | 25 | 25 | 25 | -2.62 | 27.7 | -14.93 |
| MCL-1 | 6NE5 | 1 | 25 | 25 | 25 | -1.15 | -6.99 | 88.96 |
| BFL-1 | 5UUP | 1 | 25 | 23 | 21 | -9.62 | 8.47 | -6.18 |
| BCL-B | 4B4S | 1 | 26 | 20 | 25 | -11.18 | 16.97 | 11.68 |
| TRPV1 | 6L93 | 1 | 20 | 18 | 23 | 45.11 | -11.28 | 33.63 |
| mTor | 4JT6 | 1 | 25 | 25 | 25 | 0.61 | -29.85 | -46.66 |
Table 5: Estimated Free Energy (∆ G) of each selected protein with Piperlongumine and number of interactions.
| Protein name | ∆ G (Kcal/mol) | Hydrogen bond | Pi-pi | Pi-alkyl |
|---|---|---|---|---|
| BAX | -6.1 | Arg109, Tyr164 | - | Val177, Val180, Ala183 |
| COX-2 | -8.96 | Asn368 | His372 | His193, His200, Val433 |
| caspase-9 | -4.4 | Leu384 | - | Ala388, Pro401, Ile403 |
| β-catenine | -5.8 | Ser425, Arg469 | - | Arg386, Cys429, His470 |
| JAK1 | -7.6 | Arg1007 | - | Phe958, Leu959, Leu1010 |
| JAK2 | -7.4 | Gly858 | - | Val863, Ala880, Arg980 |
| STAT3 | -6.3 | Cys108 | - | Leu21, Arg103, Arg107 |
| GSK3 | -6 | Ser203 | - | Arg220, Tyr221 |
| MCL-1 | -7.7 | Val253 | - | Phe254, Leu290, Leu246 |
| BFL-1 | -5.4 | - | - | Val44, Val48, Leu52, Val74 |
| BCL-B | -6 | Thr82 | - | Arg44, Gly84, Arg85 |
| TRPV1 | -5.9 | Glu210 | Phr245 | Tyr199, Phe236, Leu254 |
| mTor | -5.8 | Thr2207, Arg2224 | - | Leu1900, Ala2210 |
Moreover, BAX, STAT3, GSK3, and BCL-B also showed good binding affinity >6 to <7 kcal/mol and Caspase-9, B-catenin, BFL-1, TRPV1, and mTOR protein have <6, and >4 kcal/mol binding energy. The lowest binding energy shown by COX-2 protein was selected for our further computational study.
The COX-2 minimized structure (energy=-30752.73 kcal) from the Yasara web server was used for docking calculations against Piperlongumine. The docking study has shown that Piperlongumine has a potential binding affinity with -8.96 kcal/mol. It fits well in the binding region and gets formed easily.
COX-2 Interaction analysis
The COX-2 binding region has a major tunnel region with two small groves that depict the binding region as having a large area. It might fill with large molecules if you go for a pocket fitting. The COX-2 with Piperlongumine complex formed one conventional hydrogen bond with Asn368 while two non-conventional hydrogen bonds were also formed by His196, and Phe196. His193 and His372 also formed π-π stacking with the benzene ring of Piperlongumine. Similarly, His200, Val277, Leu280 and His433 were involved in π-alkyl interactions with >3Å distances. His374 involved in the formation of a non-conventional hydrogen bond and π-cation interaction with a hydroxyl group and benzene ring of Piperlongumine. The all hydroxyl group present in Piperlongumine has formed interactions with the COX-2 binding region. The compact fitting and tight interactions showed that Piperlongumine perfectly binds with the binding region of COX-2. The piperlongumine might have more than two alternate binding poses as the binding region has a large area. So, based on this hypothesis, we analyzed all 10 poses that were obtained from docking results. The alternate docked pose analysis showed that one pose has the same binding affinity as the first pose and has a slight difference in the binding region. To measure the binding pose conformations and stability of the complex, the docked complex was subjected to a 100 ns molecular dynamics simulation study shown in Figure 2.
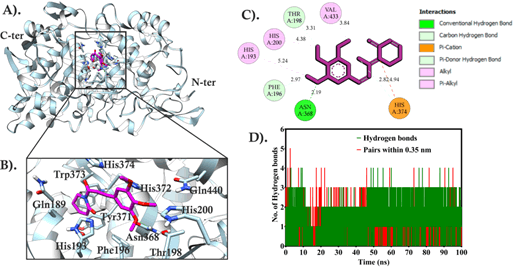
Figure 2: The molecular docking analysis of Piperlongumine with COX-2 A) COX-2 docked complex with Piperlongumine, B) Piperlongumine interacting region with COX-2 in 3D, C) Piperlongumine interacting region in 2D, and D) The number of hydrogen bonds formed by Piperlongumine within the binding region of COX-2 during 100ns simulation time.
Molecular dynamics simulation
The COX-2 in Apo form was simulated first for 100 ns simulation time with water solvent under predefined conditions. The compactness of COX-2 structure was analyzed using the radius of gyration (Rg) which showed that COX-2 maintained compactness of structure between 2.46 nm-2.53 nm till the end of the simulation time. A similar pattern adopted by the COX-2 complex maintained and slightly lowered Rg value between 2.45 nm-2.51 nm. The lowered Rg value of the COX-2 complex suggests that the piperlongumine maintained its original binding pose and not hampered the structure of COX-2 in terms of secondary structure. Similar trends were seen in RMSD values of Apo COX-2 and COX-2 complexes with piperlongumine. The Apo COX-2, RMSD has remained constant at 0.28 nm with slight fluctuations after 60 ns and maintained between 0.23 nm-0.28 nm until the simulation’s conclusion. The COX-2 complex with piperlongumine, RMSD also has a constant range of RMSD at 0.28 nm till 65 ns, after that, slightly increases and is constant at 0.31 nm maintained till the end of the simulation time. Overall, both systems have the same range of RMSD except slightly higher in the COX-2 complex, suggesting that piperlongumine has maintained the stability of the structure during the whole simulation time. The fluctuation in the local region of the COX-2 structure was analyzed using RMSF during the whole simulation time. The Apo COX-2 showed slightly higher values of RMSD fluctuating up to 0.29 nm as compared to the COX-2 complex that fluctuates up to 0.26 nm which suggests the Piperlongumine minimized the fluctuations and more stabilized the COX-2 structure during simulation. The N-terminal region (18 aa-250 aa) showed many high peaks fluctuating in both Apo as well as in the Complex form of COX-2 due to the large loop region present in this region while the C-terminal region (258 aa-564 aa) has helical and sheet region that stabilized the structure with stable values of RMSF up to 0.12 nm. The COX-2 complex with Piperlongumine has lowered RMSF values compared to Apo suggests that Piperlongumine has been able to minimize the fluctuations in the loop region present in the N-terminal region.
The piperlongumine heavy atoms RMSD started with a lowered range of RMSD between 0.09 nm-0.12 nm till 10 ns, after 12 ns, RMSD suddenly changed at 0.21 nm and kept being the same till the closing stages of the simulation time. This sudden change might be due to changes in the binding mode of Piperlongumine in the binding region and this change causes the most stable form of Piperlongumine to be maintained till the end of the simulation time. Overall, our observation suggests that piperlongumine has been able to stabilize the COX-2 in a given simulation time and can inhibit the function of COX-2 shown in Figure 3.
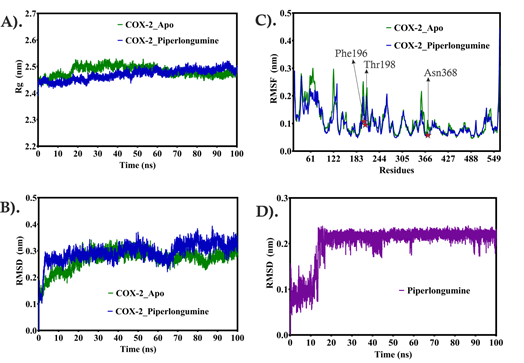
Figure 3: The molecular dynamics simulation study of Apo and Complex form of COX-2. A) COX-2 structure compactness of Apo and in the presence of Piperlongumine, B) Stability analysis of Apo COX-2 and in the presence of Piperlongumine, C) Fluctuations analysis in the local region in Apo and in the presence of Piperlongumine, and D) Piperlongumine stability during 100 ns simulation time.
Free energy analysis
The Piperlongumine has lowest binding free energy with 59.01 kcal/mol and in comparison with complex form it has -497.31 kcal/mol. The COX-2 alone has -485.75 kcal/mol while complex form lowered with -497.31 kcal. This indicates the piperlongumine is able to attain the lowest free binding energy during simulation shown in Table 6.
Table 6: Total calculated binding free energies of Piperlongumine with COX-2 complex.
| Energy Component kcal/mole | Ligand | Receptor | Complex | Difference | ||||
|---|---|---|---|---|---|---|---|---|
| Average | Std. Dev | Average | Std. Dev | Average | Std. Dev | Average | Std. Dev | |
| Δ Evdw | -6.46 | 0.41 | -211.71 | 24.1 | -220.57 | 26.11 | 90.86 | 3.21 |
| Δ Eele | -17.11 | 0.89 | -174.8 | 88.4 | -182.69 | 91.21 | -10.63 | 4.17 |
| Δ EGB | -7.43 | 0.34 | -541.34 | 71.07 | -551.21 | 73.47 | 27.37 | 3.47 |
| Δ E SURF | 1.09 | 0.11 | 88.09 | 2.41 | 89.29 | 3.07 | -6.52 | 1.42 |
| Δ G Gas | 55.21 | 2.72 | -108.21 | 102.32 | -112.37 | 104.61 | -59.91 | 4.14 |
| Δ G sol | -3.03 | 0.66 | -425.54 | 81.46 | -428.01 | 84.29 | 20.84 | 3.12 |
| Δ G total | 59.01 | 3.01 | -485.75 | 63.92 | -497.31 | 65.17 | -39.07 | 3.82 |
Discussions
Unquestionably, colorectal cancer is a common and incapacitating condition marked by chronic changes in bowel habits, such as constipation, diarrhoea, or changes in faecal consistency Traditional medicines can only briefly relieve clinical symptoms; they are ineffective at curing colorectal cancer. Therefore, the need for more efficient treatment options in order to slow the spread of colorectal cancer is very critical. Against a variety and wide spectrum of cancers, piperlongumine has demonstrated strong action and promising therapeutic prospects. We therefore looked into the putative mechanisms underpinning piperlongumine’s protective action against the onset of colorectal cancer.
Our study came out with few concrete findings which are also confirmed by other research articles which includes, presence of a hydrophobicity region indicating that piperlongumine can bind to the tunnel-like region which is present in the protein structure. Our docking calculation showed that COX-2, JAK1, JAK2, and MCL-1 proteins have the lowest binding free energy compared to other proteins similar to few other studies. Piperlongumine has been able to stabilize the COX-2 in a given time simulation and can inhibit the function of COX-2. In our study, similar to other scientific research findings, piperlongumine was able to attain the lowest free binding energy during simulation. Looked into whether piperlongumine has anti-inflammatory and anti-amyloidogenic effects by suppressing NF-B. LPS (0.25 mg/kg/day, i.p.) was administered intraperitoneally (i.p.) to create a mouse model of LPS-induced memory impairment. According to the outcomes of their behavioural testing, piperlongumine decreased LPS-induced memory. Piperlongumine also reduced the actions of secretases and stopped beta-amyloid (A) build up brought on by LPS. These results were connected to the suppression of NF-B activation. Piperlongumine binds to p50, as demonstrated by a pull-down test and examination of docking models. Their research suggests that piperlongumine inhibits NF-B signalling to reduce LPS-induced amyloidogenesis and neuroinflammation, which suggests that piperlongumine, may be used to treat Alzheimer’s disease looked into the chemical and physical interactions of piperlongumine with anti-inflammatory substances, as well as their impact on cell migration and proliferation, as well as the gene expression of inflammatory mediators. Piperlongumine may interact with a peptide of annexin A1 (ANXA1), an endogenous anti-inflammatory mediator with therapeutic promise in cancer, according to molecular docking data and physicochemical studies. Self-organising maps were used by Jao Conde to deconvolute the phenotypic effects of piperlongumine and establish a connection to the modification of the human transient receptor potential vanilloid 2 (hTRPV2) channels. In order to learn more about the structures of eight natural product-inspired analogues, were inspired by the amazing bioactivities displayed by the natural product piperlongumine. Through the use of two-dimensional (2D) 15-N-based spectroscopy, nuclear magnetic resonance (NMR) spectroscopy, and other methods, they were able to confirm the structure of the important cyclised dihydropyrazole carbothioamide piperine analogues for the first time. Prior studies showed promising outcomes from these scaffolds for the suppression of the IL-1 and NF-B pathway-mediated inflammatory response. However, it is still unclear how these chemicals interact with the proteins that are their targets on a molecular level. Recent publications in peer-reviewed journals have shown a number of different pathways by which the pyridine alkaloid piperlongumine causes a range of anticancer actions. A novel mechanism by which this alkaloid mediates its anticancer effects was demonstrated. Piperlongumine-Chemosensitizes- Tumor-Cells-through piperlongumine blocked NF-B activated by TNF and various other cancer promoters. Along with this downregulation, IB’s phosphorylation and degradation were inhibited. This pyridine alkaloid directly interacts with IKK and suppresses its function, according to further research. IKK was inhibited by interacting with its cysteine 179, as piperlongumine’s activity was eliminated when this residue was changed to an alanine. The expression of proteins involved in cell survival (Bcl-2, Bcl-xL, c-IAP-1, c-IAP-2, survivin), proliferation (c-Myc, cyclin D1), inflammation (COX-2, IL6), and invasion (COX-2, IL6) was downregulated when NF-B activity was inhibited (ICAM-1, -9, CXCR-4, VEGF). Overall, their findings point to an unique mechanism by which down regulating proinflammatory pathways allows piperlongumine to demonstrate anticancer action.
Despite improvements in the creation of diagnostic and therapeutic techniques, CRC (colorectal cancer) continues to be a significant cause of cancer-related death. It has been shown that PL mediates effects via causing ROS-dependent ER stress and mitochondrial dysfunction in CRC, as stated in Reactive oxygen species (ROS) levels and antioxidant activity are higher in malignant cells than in normal ones. They are therefore more susceptible to substances that raise intracellular ROS since they have greater baseline ROS levels. Previous studies have also demonstrated that the synergistic cytotoxicity brought on by combination treatments is unique to cancer cells. In some earlier research, PL was discovered to be a direct TrxR1 inhibitor with anti-cancer properties piperlongumine as a direct TrxR1 inhibitor with suppressive activity against gastric cancer.
In an earlier research that was conducted. It has been discovered that PL caused an excessive amount of ROS to be produced because it depleted glutathione and inhibited thioredoxin reductase. Second, PL increased the intrinsic and hypoxic radiosensitivity of tumour cells, which was connected to an increase in DNA damage brought on by ROS, a halt in the G2/M cell cycle, and a suppression of cellular respiration. Additionally, the radiosensitizing effects of PL were confirmed in vivo by the researchers. They ultimately came to the conclusion that PL could function as a radiosensitizer in colorectal cancer. A previous study has shown that PL is effective against CRC in both in vitro and in vivo settings. According to one such study, PL generated cytotoxicity on CRC cells primarily by suppressing cyclin D1, which was responsible for preserving the tumorigenicity of the CRC cells. PL also inhibited the levels of c-Fos in CRC cells through adversely regulating the Akt and ERK1/2 signalling pathways. The same group has shown in another in vivo study that PL considerably slowed the growth of the tumour in the xenograft mice model of CRC. Additionally, it has been shown in a research that PL suppressed mitogen- activated protein kinase (MAPK)/ERK kinase (MEK) signaling in CRC cells, leading to dose and time-dependent cell death. It was also shown that PL significantly decreased the development of the CRC cell line SW-620. Additionally, it was demonstrated that PL blocked the JNK signalling pathway, causing CRC cells to undergo apoptosis. Additionally, without affecting the expression of Bax, p21, or p53, this substance was able to cause cytotoxicity and apoptosis in CRC cells. It has also been shown that PL targets the TrxR and GSH anti-oxidant systems and induces ROS production in CRC cells. Additionally, increased ROS production by PL caused DNA damage and cell cycle arrest in the CRC cells. Additionally, it was discovered that this substance caused the restoration of wild-type p53 function and reduced tumour development in a nude mice model. Furthermore, in the azoxymethane (AOM)/DSS-induced animal model of CRC, treatment of PL may drastically reduce levels of COX-2, IL-6, -catenin, and snail, hence reducing inflammation and tumour growth. Additionally, it was shown that PL caused morphological modifications and nuclear damage in CRC, which accelerated apoptosis and cell death. In addition, PL markedly raised the amounts of ROS within cells, which also caused apoptosis, as shown by the alteration of the expression of many proteins related to the process, including Bax, Bcl-2, survivin, p53, and p21. Additionally, it was discovered that PL reduced the CRC cell’s ability to migrate. The main barrier to employing the existing conventional chemotherapeutic drugs for cancer is chemoresistance, Numerous researches have examined the potential of PL as a strong and reasonably priced anti-cancer medication throughout the years. The essential elements of the crucial signalling pathways that play a role in the development of chemoresistance in cancer cells were discovered to be modulated by PL. As a result, current efforts have concentrated on creating PL as a chemosensitizer that made cancer cells more susceptible to commercially available chemotherapeutics. The function of PL in making cancer cells more susceptible to various crucial medications is discussed in the section that follows. The development of PL as a chemosensitizer and radiosensitizer, which sensitised cancer cells to commercially available chemotherapeutics such cisplatin, doxorubicin, 5-FU, gemcitabine, oxaliplatin, and PTX, as well as ionization/X-ray radiation, has recently received significant attention. PL inhibits the biotransformation of the medications into less active metabolites, suppresses the characteristics of cancer stem cells, and acts on the cell signaling pathways and genes related with chemo and radioresistance to sensitise the cancer cells to chemotherapeutics and radiation. A very low concentration of PL was also discovered to have anti-cancer properties, which tends to lessen the possibility of toxicity to nearby normal cells and organs as well as negative side effects. In actuality, PL was said to have cardioprotective and hepatoprotective properties. In order to somewhat alleviate the negative effects brought on by the traditional chemotherapeutics, this chemical may be employed as an adjuvant.
The hydrophobic property of PL, which contributes to its limited bioavailability, is the fundamental barrier to its development as a conventional chemotherapy. However, studies show that when administered in conjunction, PL increased the bioavailability of docetaxel and so worked as a bio enhancer. In order to increase the bioavailability of PL, drug carriers such as hydrogels, liposomes, microspheres, and nanoparticle formulations can be modified. Additionally, it was discovered that the pharmacokinetic profile of the PL-loaded nanoemulsion was superior to that of the drug’s pure form.
Although PL has demonstrated enormous promise in the prevention and treatment of several malignancies, more research is necessary before PL can be developed into a clinical chemotherapeutic agent. These investigations are included below:
1. A variety of experimental cancer models should be used to assess the chemopreventive effects of PL.
2. Research on the substance’s cytotoxicity in various organs should be done.
3. Molecular indicators need to be created to assess the effectiveness of PL in multicenter clinical studies that are randomised.
4. Comprehensive research in people is needed to determine the bioavailability of PL and its metabolic and toxicological profile.
5. PL effective bioformulations intended for prolonged release should be created.
Due consideration should thus be given to carrying out such investigations on this molecule in order to turn it into a possible anti-cancer medication. It is also important to note the fact that PL is safe and makes it easier to assess its therapeutic benefits in future clinical studies.
Study on cytotoxicity has shown that PL inhibits INT-407 and HCT-116 cell growth in a concentration and time-dependent manner. The quantities of intracellular reactive oxygen species were also increased by PL, which may cause fatal oxidative stress, mitochondrial malfunction, and nuclear fragmentation. Surprisingly, P53, P21, BAX, and SMAD4 were considerably elevated, but BCL2 and SURVIVIN were dramatically down regulated, following PL therapy. These results imply that PL inhibits the growth of intestinal cancer cells. It is crucial that these natural compounds undergo clinical assessment through a significant number of clinical studies in order to confirm the efficacy shown in vitro and in animal models as more information is obtained in the design of innovative drug delivery vehicles. Additionally, it’s important to determine each compound’s safety profile before using it on cancer patients. Despite the challenges that still need to be overcome; the immense potential of these bioactive substances produced from plants makes it worthwhile for the scientific community to work together for the benefit of oncological patients.
Conclusion
By prioritising biochemical screens, statistical learning helps speed up the drug development process. These techniques should work for both manufactured and natural compounds. This idea implies that our computer-driven, disease-neutral methodology is a workable way to swiftly repurpose chemical matter and support the creation of novel therapies. Through molecular docking with numerous colon cancer target proteins, the mechanism of action of piperlongumine was defined the efficiency and frequency of interactions between piperlongumine and amino acids. We collectively demonstrated the ability of computers to perceive complex data patterns and make decisions based on them, supported by functional and structural data. We anticipate this capability will play a larger role in the future development of new drugs as well as the effectiveness of colorectal cancer treatments. This study promotes the use of piperlongumine-containing compounds as potential colorectal cancer therapy. This lead chemical will be valuable to the scientific community in the pursuit of novel colorectal cancer treatments.
Authors Contributions
IS and AK conceived the study. All experiments were designed by IS and AK, PK. All experiments were carried out by IS, AK, PK and HPM. Data analyses were by IS and HPM. Work was supervised by AK Manuscript was prepared by IS and reviewed by IS, HPM, PK and AK. All authors read and approved the final manuscript.
Acknowledgement
All the authors are thankful to CSIR and the Department of Biotechnology, Faculty of Engineering and Technology, Rama University, Kanpur, Uttar Pradesh (India).
Conflict of Interest
There are no conflicts of interest among the authors of this study.
References
- Jemal, T. Murray, A. Samuels, A. Ghafoor, E. Ward, et al. CA: A Cancer Journal for Clinicians, CA Cancer J Clin, 53(2003): 5-26.
- C.E. Eberhart, R.J. Coffey, A. Radhika, F.M. Giardiello, S. Ferrenbach, et al. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas, Gastroenterology, 107(1994): 1183-1188.
- R.N. DuBois, A. Radhika, B.S. Reddy, A.J. Entingh, Increased cyclooxygenase-2 levels in carcinogen-induced rat colonic tumors, Gastroenterology, 110(1996): 1259-1262.
- C.S. Williams, C. Luongo, A. Radhika, T. Zhang, L. Lamps, et al. Elevated cyclooxygenase-2 levels in Min mouse adenomas, Gastroenterology, 111(1996): 1134-1140.
- G.M. Denicola, F.A. Karreth, T.J. Humpton, A. Gopinathan, C. Wei, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis, Nature, 475(2011): 106-110.
- M. Jeanne, V. Lallemand-Breitenbach, O. Ferhi, M. Koken, M. Le, et al. Thé PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3, Cancer Cell, 18(2010): 88-98.
- J. Alexandre, Y. Hu, W. Lu, H. Pelicano, V. Huang, Novel action of paclitaxel against cancer cells: Bystander effect mediated by reactive oxygen species Cancer Res, Cancer Res, 67(2007): 3512-3517.
- J. Saczko, A. Choromańska, N. Rembiałkowska, M. Dubińska-Magiera, I. Bednarz-Misa, et al. Oxidative modification induced by photodynamic therapy with Photofrin®II and 2-methoxyestradiol in human ovarian clear carcinoma (OvBH-1) and human breast adenocarcinoma (MCF-7) cells, Biomed Pharmacother, 71(2015): 30-36.
- M.J. Thun, M.M. Namboodiri, C.W. Heath, Aspirin use and reduced risk of fatal colon cancer, N Engl J Med, 325(1991): 1593-1596.
- E. Giovannucci, K.M. Egan, D.J. Hunter, M.J. Stampfer, G.A. Colditz, Aspirin and the risk of colorectal cancer in women, N Engl J Med, 333(1995): 609-614.
- E. Giovannucci, E.B. Rimm, M.J. Stampfer, G.A. Colditz, A. Ascherio, et al. Aspirin use and the risk for colorectal cancer and adenoma in male health professionals, Ann Intern Med, 121(1994): 241-246.
- W.L. Smith, D.L. DeWitt, R.M. Garavito, Cyclooxygenases: Structural, cellular, and molecular biology, Annu Rev Biochem, 69(2000): 145-182.
- H.R. Herschman, W. Xie, S. Reddy, Inflammation, reproduction, cancer and all that… The regulation and role of the inducible prostaglandin synthase, Bioessays, 17(1995): 1031-1037.
- H.R. Herschman, Prostaglandin synthase 2, Biochim Biophys Acta, 1299(1996): 125-140.
- W.L. Xie, J.G. Chipman, D.L. Robertson, R.L. Erikson, D.L. Simmons, Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing, Proc Natl Acad Sci USA, 88(1991): 2692-2696.
- D.A. Kujubu, B.S. Fletcher, B.C. Varnum, R.W. Lim, H.R. Herschman, TIS10, a phorbol ester tumor promoter- inducible mRNA from Swiss 3T3 cells, encodes a novel prostaglandin synthase/cyclooxygenase homologue, J Biol Chem, 266(1991): 12866-12872.
[Google Scholar] [PubMed]
- T. Hla, K. Neilson, Human cyclooxygenase-2 cDNA, Proc Natl Acad Sci USA, 89(1992): 7384-7388.
- R.A. Soslow, A.J. Dannenberg, D. Rush, B.M. Woerner, K.N. Khan, et al. COX-2 is expressed in human pulmonary, colonic, and mammary tumors, Cancer, 89(2000): 2637-2645.
- G.A. FitzGerald, C. Patrono, The coxibs, selective inhibitors of cyclooxygenase-2, N Engl J Med, 345(2001): 433-442.
- L.J. Marnett, A.S. Kalgutkar, Cyclooxygenase 2 inhibitors: Discovery, selectivity and the future, Trends Pharmacol Sci, 20(1999): 465-469.
- F.A. Sinicrope, M. Lemoine, L. Xi, P. Lynch, K.R. Cleary, et al. Reduced expression of cyclooxygenase 2 proteins in hereditary nonpolyposis colorectal cancers relative to sporadic cancers, Gastroenterology, 117(1999): 350-358.
- H. Sano, Y. Kawahito, R.L. Wilder, A. Hashiramoto, S. Mukai, et al. Expression of cyclooxygenase -1 and -2 in human colorectal cancer, Cancer Res, 55(1995): 3785-3789.
[Google Scholar] [PubMed]
Copyright: © 2023 Indrajeet Singh, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.