Research Article - Journal of Drug and Alcohol Research ( 2022) Volume 11, Issue 1
Structure Based Virtual Screening of Selected Phytochemicals aganist C1156y Mutant Anaplastic Lymphoma Kinase (ALK) using Molecular Docking and Admet Prediction
Kanaka Durga Devi N1*, Bharghava Bhushan Rao P2, Ramya B3, Sireesha U3, Chandana M3, Bhavana K3, Likitha N3, Harisitha P3 and Ravi Sankar K32Department of Pharmaceutics, V V Institute of Pharmaceutical Sciences, India
3Department of Pharmacy Practice, Siddhartha College of Pharmaceutical Sciences, India
Kanaka Durga Devi N, Department of Pharmaceutics and Biotechnology, KVSR Siddhartha College of Pharmaceutical Sciences, India, Email: Deviriss@gmail.com
Received: 31-Dec-2021, Manuscript No. jdar-22-52015; Editor assigned: 05-Jan-2022, Pre QC No. jdar-22-52015 (PQ); Reviewed: 19-Jan-2022, QC No. jdar-22-52015; Revised: 24-Jan-2022, Manuscript No. jdar-22-52015 (R); Published: 31-Jan-2022, DOI: 10.4303/jdar/236156
Abstract
Lung cancer is one of the fatal malignancies in both men and women worldwide. Anaplastic Lymphoma Kinase (ALK) rearrangements triggered the fusion of echinoderm microtubule-associated protein-like 4 (EML4) and ALK increasing the tyrosine kinase activity leading to cell proliferation and tumourigenesis causing Non-Small Cell Lung Cancer (NSCLC). Crizotinib emerged as a first-generation ALK inhibitor; however it developed resistance within a year of initiation of treatment in a few patients with deteriorating therapeutic outcomes. Various secondary mutations in ALK are responsible for crizotinib resistance in patients. C1156Y is a point mutation seen in patients resistant to crizotinib. Although second and third-generation ALK inhibitors are currently available, there is still a need to develop new ALK inhibitors as the existing compounds are developing resistance. In this study, we screened 940 phytochemicals against C1156Y mutant ALK to identify binding affinities. The results showed that three steroidal alkaloids Conessine, Solanocapsine, and Muldamine exhibited binding affinity (-9.0 kcal/mol) and are predicted to show activity in CNS for the prevention and management of CNS metastasis. We conclude that these 3 steroidal alkaloids are worth further in vitro and in vivo studies to emerge as potent C1156Y mutant ALK inhibitors.Keywords
C1156Y mutant ALK; Phytochemicals; Molecular docking; Structure based screening
Introduction
Lung cancer is one of the deadliest malignancies accounting for 1.5 million deaths worldwide. Lung cancer can be broadly divided based on histology into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC accounts for almost 80-85% of the lung cancer cases [1]. Anaplastic Lymphoma Kinase (ALK) rearrangements are seen in 5% of NSCLC patients, equivalent to over 60,000 patients diagnosed annually worldwide [2]. Apart from NSCLC, ALK is also found in anaplastic large cell lymphoma, neuroblastoma and inflammatory myofibroblastic tumour [3]. ALK gene is located on chromosome 2 (2p23). It encodes for ALK receptor tyrosine kinase which belongs to insulin receptor super family [4]. In NSCLC, ALK rearrangements lead to fusion of echinoderm microtubule- associated protein-like 4 (EML4) and ALK further lead to the activation of kinase activity [5]. So, ALK inhibitors emerged as best therapeutic agents against ALK rearranged NSCLC. Crizotinib belongs to the first generation ALK inhibitors with outstanding efficacy and safety. However resistance to crizotinib developed within one or two years of treatment initiation [6].
Point mutations in ALK lead to crizotinib drug resistance. Many secondary mutations like C1156Y, L1196M etc. are identified within the kinase domain of EML4-ALK. C1156 residue of ALK is located at the non-active site and thus has no direct interaction with crizotinib;however, C1156Y displaces crizotinib and affects the conformations of loop 1122–1131, b-sheet 1145–1152, and a-helix 1157–1174 [7]. Although second and third generation ALK inhibitors are present, there is still an immense need of ALK inhibitors due to different mutations in ALK. So, in this study we have focused on phytochemicals as they have reported various pharmacological activities.
Method and Methodology
Preparation of protein
The X-Ray Diffraction structure of C1156Y Mutant ALK (PDB ID:2YJS) with resolution 1.9Ao was retrieved from Protein Data Bank (PDB). The hetero atoms present in the crude PDB file were removed. Kollman charges and polar hydrogen atoms are added to satisfy their appropriate charges in Autodock Vina and the file is converted to PDBQT format [8].
Identification of Binding Pockets
Active pockets in 2YJS receptor molecule were predicted using Computed Atlas of Surface Topography of Proteins (CASTp) program[9] (http://sts.bioe.uic.edu/castp/). One binding pocket was predicted with solvent accessible surface area and volume of 633.8 and 1019.2 respectively. These catalytic sites are further used for identifying ligands with best affinity to the receptor.
Preparation of Ligands
The phytochemicals (940) along with Standard ALK inhibitors viz. Ceritinib, Alectinib, Brigatinib, Crizotinib and Lorlatinib were considered for molecular docking study. 3D structures of ligands were retrieved from Pub Chem compound database at NCBI (https://pubchem.ncbi.nlm. nih.gov/). Ligands downloaded in SDF format are coverted to PDB format using PyMOL (https://pymol.org/2/). In Auto Dock Vina, gastgeir charges and hydrogens were added and converted to PDBQT format.
Molecular Docking
Following protein and ligand preparation, molecular docking analysis was performed using Autodock Vina to assess their binding affinities. To define the binding site a grid box was set at 44 × 48 × 16 A0 along the X, Y and Z axes respectively at grid resolution of 0.4 A0. The various conformations for ligand in docking procedure were generated. The dock score of the best poses docked into the receptor for all the ligands was considered. The docking scores of standard ALK inhibitors were compared with phytochemicals to identify the phytochemicals with best affinity to mutated ALK receptor (Figure 1).
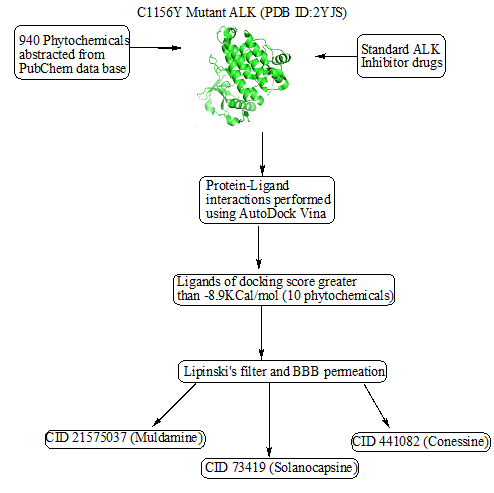
Figure 1: Flow chart of structure based screening of phytochemicals against mutant ALK.
ADME and Druglikeliness
Subsequent to molecular docking, phytochemicals with higher affinity to receptor than standard ALK inhibitors were further studied for prediction of Absorption, Distribution, Metabolism, Excretion (ADME) parameters and drug likeliness in SwissADME web tool [10](http://www.swissadme. ch/). Predicted parameters like Molecular weight, Numer of H-bond acceptors, H-bond donors, LogPo/w (Lipophilicity), LogS (solubility), GI absorption, Blood brain barrier(BBB) permeation were recorded. Lipinski’s filter was applied for examining drug likeliness.
Results and Discussion
Initially docking was carried for standard ALK inhibitors and best binding affinities were exhibited by Alectinib (-8.9 kcal/mol) and Lorlatinib (-8.9 kcal/mol) as shown in Table 1 and low affinity was exhibited by crizotinib clearly explaining that C1156Y mutation significantly affects its binding to ALK.
Table 1: Structures and Binding affinities of standard ALK inhibitors.
Standard Alk Inhibitors |
Binding affinity (Kcal/mol) | Structures |
|---|---|---|
| 1. Ceritinib | -7.9 |
|
| 2. Alectinib | -8.9 |
|
| 3. Brigatinib | -8.5 |
|
| 4. Crizotinib | -7.8 |
|
| 5. Lorlatinib | -8.9 |
|
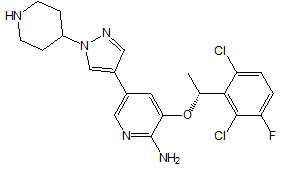
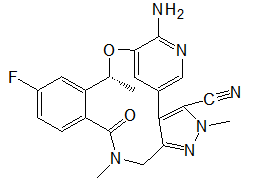
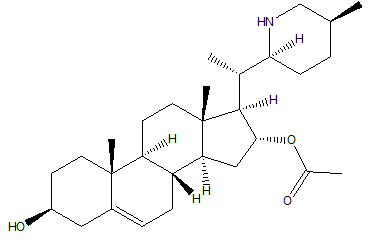
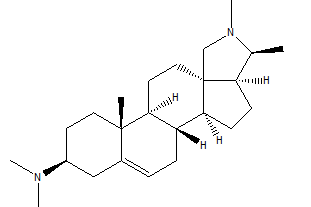
The 940 phytochemicals were also docked and 10 phytocheMicals exhibited dock score greater than (-8.9 kcal/ mol).These were screened for further investigation as they exhibited greater binding affinities than Alectinib and Lorlatinib Table 2. CID 11316166 (Cavicularin) exhibited the highest affinity of -9.9 Kcal/mol followed by CID 5315779 (Cepharanoloine) and CID 5492110 (Ochnaflavone) of binding affinity -9.8 kcal/mol each. Alecitinib and Lorlatinib showed interaction with ASP 1270, MET 1199, LYS 1150, ARG 1253, ASP 1203.Whereas the shortlisted phytochemicals interacted with ASP 1270, MET 1199, ASP 1203,ARG 1275, GLY 1269,GLY 1123 showing that they are binding to catalytic residues.
Table 2: Binding affinities, Pharmacokinetic parameters and druglikeliness of shortlisted phytochemicals.
| Phytochemicals | Binding Affinity (kcal/mol) | Molecular weight (g/mol) |
Number of H-bond acceptors | Number of H-bond donors | Log Po/w | Log S | GI absorbtion | BBB permeant | Lipinski filter |
|---|---|---|---|---|---|---|---|---|---|
| CID 11316166 | -9.9 | 422.4 | 4 | 3 | 5.06 | -7.98 | High | No | Yes; 1 violation |
| CID 5315779 | -9.8 | 592.6 | 8 | 1 | 5.00 | -7.76 | High | No | Yes; 1 violation |
| CID 5492110 | -9.8 | 538.4 | 10 | 5 | 4.01 | -7.14 | Low | No | Yes; 1 violation |
| CID 10206 | -9.4 | 606.71 | 8 | 0 | 5.36 | -7.98 | High | No | Yes; 1 violation |
| CID 76972186 | -9.4 | 598.6 | 8 | 2 | 4.51 | -6.09 | High | No | Yes; 1 violation |
| CID 15922818 | -9.3 | 611.53 | 14 | 9 | -0.07 | -3.70 | Low | No | No; 3 violations |
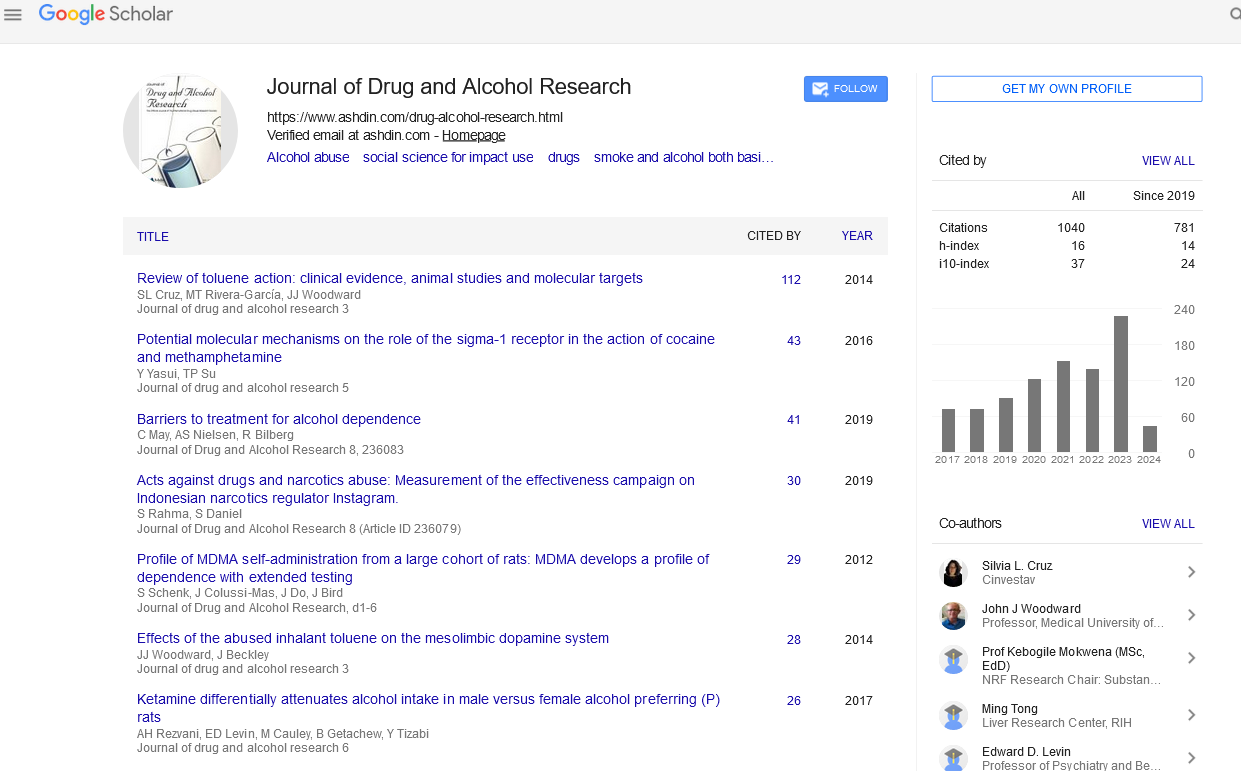
| CID 442105 | -9.2 | 518.70 | 3 | 3 | 3.75 | -6.88 | High | Yes | No; 2 violations |
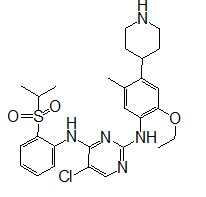
| CID 73419 | -9.0 | 430.67 | 4 | 3 | 4.00 | -5.65 | High | Yes | Yes; 1 violation |
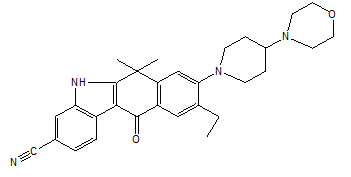
| CID 441082 | -9.0 | 356.59 | 2 | 0 | 4.36 | -5.04 | High | Yes | Yes; 1 violation |
| CID 21575037 | -9.0 | 518.70 | 3 | 3 | 3.75 | -6.88 | High | Yes | Yes; 1 violation |
CNS involvement is seen in advanced ALK rearranged NSCLC. The major therapeutic issue in ALK rearranged NSCLC patients is control and prevention of CNS metastasis [2]. Crizotinib failed to show promising outcomes in controlling CNS metastasis. However the second generation ALK inhibitors v.i.z. Alectinib and brigatinib were active for CNS metastasis [11]. So, it is desired that the phytochemical of choice shows its activity in CNS as well by BBB permeation. Among the shortlisted phytochemicals only 3 were predicted to cross BBB and showed only 1 violation in lipinski’s rule of five revealing favorable drug-like properties Table 3.
Table 3: Structure and Binding affinities of Steroidal alkaloids.
| Phytochemicals | Binding affinity (Kcal/mol) | Structures |
|---|---|---|
| CID 21575037 | -9.0 |
|
| CID 441082 | -9.0 |
|
| CID 73419 | -9.0 |
|
These three phytochemicals are steroidal alkaloids with a perhydro-1, 2-cyclopentanophenanthrene nucleus. Steroidal alkaloids are the natural nitrogen-containing compounds that are present in many edible or medicinal plants. Steroidal alkaloids are studied extensively and showed to have anti-microbial, anti-nociceptive, anti-inflammation and anticancer properties [12].
Conessine is a steroid alkaloid derived from many plants of family Apocynaceae. Conessine is well known as a Histamine antagonist selective to H3 subtype with high BBB penetration and is also found to have long CNS clearance time [13]. It’s high affinity to C1156Y mutant ALK and high BBB permeation reveals conessine can be studied further and may emerge as a novel phytochemical for ALK rearranged NSCLC patients with C1156Y mutation. Solanocapsine is derived from Solanum pseudocapsicum. It is studied previously for its acetylcholine esterase inhibition activity [14]. Muldamine, a phytosterol alkaloid was extracted from the roots and rhizomes of Veratrum californicum [15]. Muldamine is unexplored for its biological activity. So, there is a need to highlight these steroidal alkaloids for emerging as anti-oncogenic drugs against ALK rearranged NSCLC.
Conclusion
In the present study we screened various phytochemicals against C1156Y mutant ALK. Highest affinity was shown by CID 11316166. However considering BBB permeation three steroidal alkaloids CID 73419, CID 441082, CID 21575037 showed affinity to mutant ALK. We conclude that further in vitro and in vivo studies should be performed to assess the therapeutic potential of these three steroidal alkaloids against mutant ALK rearranged NSCLC.
Acknowledgements
The authors are thankful to the management and principal of KVSR Siddhartha College of Pharmaceutical Sciences, Vijayawada, Andhra Pradesh, India.
Conflicts of Interest
We hereby declare that the authors have no conflicts of interest
References
- K. Ramanathan, S. Maiti, V. Shanthi, W. Shin, D. Kihara, Implementation of pharmacophore-based 3D QSAR model and scaffold analysis in order to excavate pristine ALK inhibitors, Med Chem Res, 28 (2019), 1726–1739.
- M. Thomas, K. J. O. Byrne, D. Moro-sibilot, D. R. Camidge, T. Mok, et al., Crizotinib versus chemotherapy in advanced, N Engl J Med, 25 (2015) 2385–2394.
- D. Fontana, M. Ceccon, C. Gambacorti-passerini, L. Mologni, Activity of second-generation ALK inhibitors against crizotinib-resistant mutants in an NPM-ALK model compared to EML4-ALK, Cancer Med, 4 (2015), 953–965.
- J. Duyster, R. Bai , S. W. Morris, Translocations involving anaplastic lymphoma kinase (ALK), J Oncgn, 20 (2001), 5623–5637.
- Q. Huang, T. W. Johnson, S. Bailey, A. Brooun, K. D. Bunker, et al., Design of potent and selective inhibitors to overcome clinical anaplastic lymphoma kinase mutations resistant to crizotinib, J Med Chem, 57 (2014), 1170–1187.
- J. F. Gainor, S. Bergqvist, A. Brooun, B. J. Burke, Resensitization to crizotinib by the lorlatinib, N Engl J Med, 374 (2016), 54–61.
- H. Sun, F. Ji, Biochemical and biophysical research communications a molecular dynamics investigation on the crizotinib resistance mechanism of c1156y mutation in ALK, Biochem Biophys Res Commun, 423 (2012), 319–324.
- O. Trott, A. J. Olson Software, News and update autodock vina : Improving the speed and accuracy of docking with a new scoring function, J Comput Chem, 31 (2009), 455–461.
- W. Tian, C. Chen, X. Lei, J. Zhao, J. Liang, Computed atlas of surface topography of proteins, 46 (2018), 363–367.
- A. Daina, O. Michielin, V. Zoete, Swiss ADME : A free web tool to evaluate pharmacokinetics , drug- likeness and medicinal chemistry friendliness of small molecules, Nat Publ Gr, 7 (2017) 1–13.
- J. Wu, J. Savooji, D. Liu, Second- and third-generation ALK inhibitors for non-small cell lung cancer, J Hematol Oncol, 6 (2016), 1–7.
- Q. Jiang, M. Chen, K. Cheng, P. Yu, X. Wei, Therapeutic potential of steroidal alkaloids in cancer and other diseases, Med Res Rev, 36 (2015), 119–143.
- V. J. Santora, J. A. Covel, R. Hayashi, B. J. Hofilena, J. B. Ibarra, A new family of H 3 receptor antagonists based on the natural product Conessine, Bioorg Med Chem Lett, 18 (2008), 1490–1494.
- M. E. García, J. L. Borioni, V. Cavallaro, M. Puiatti, B. Pierini, Solanocapsine derivatives as potential inhibitors of acetylcholinesterase : Synthesis , Molecular Dckng Bio Strds, 104 (2015), 95–110.
- C. M. Chandler, J. W. Habig, A. Fisher, K. V. Ambrose, S. T. Jiménez, Improved Extraction and Complete Mass Spectral Characterization of Steroidal Alkaloids from veratrum californicum, Nat Prod Commun, 8 (2013), 1059–1064.